##########
# PROTEIN: Q2246_545_115.5wLII_11077_4 110 115 # Category_B
# Length: 110
# Weight: 12482.62
# Similarity: UniProt_A3ESD2, BLAST_A3ESD2, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
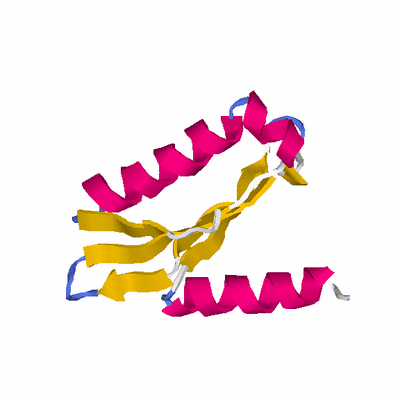
# Cat 79.09 M_01 2fck_A 181 1 4e-10 20.000 87:87 (23-109:54-140)
# Evl 72.73 M_00 2vqu_B 846 2 6e-17 13.000 80:92 (10-100:454-534)
# Sid 32.73 M_00 2p3j_A 161 1 0.020 44.000 36:36 (15-50:29-64)
# Cov 83.64 M_04 2z0z_A 194 1 2e-07 15.000 92:118 (2-110:32-132)
# LaL 79.09 M_01 2fck_A 181 1 4e-10 20.000 87:87 (23-109:54-140)
# OvN 83.64 M_04 2z0z_A 194 1 2e-07 15.000 92:118 (2-110:32-132)
# OvC 83.64 M_04 2z0z_A 194 1 2e-07 15.000 92:118 (2-110:32-132)
PDB: 2fck PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 12-DEC-05 2FCK
TITLE STRUCTURE OF A PUTATIVE RIBOSOMAL-PROTEIN-SERINE
TITLE 2 ACETYLTRANSFERASE FROM VIBRIO CHOLERAE.
SOURCE 2 ORGANISM_SCIENTIFIC: VIBRIO CHOLERAE O1 BIOVAR ELTOR STR.
SOURCE 4 ORGANISM_TAXID: 243277;
KEYWDS VIBRIO CHOLERAE, SERINE ACETYLTRANSFERASE, RIBOSOMAL-
KEYWDS 2 PROTEIN, STRUCTURAL GENOMICS, PSI, PROTEIN STRUCTURE
KEYWDS 3 INITIATIVE, MIDWEST CENTER FOR STRUCTURAL GENOMICS, MCSG,
KEYWDS 4 UNKNOWN FUNCTION
SCOP: d.108 Acyl-CoA N-acyltransferases (Nat)
SCOP: d.108.1 Acyl-CoA N-acyltransferases (Nat)
SCOP: d.108.1.1 N-acetyl transferase, NAT
PDB: 2vqu PDBsum
HEADER HYDROLASE 18-MAR-08 2VQU
TITLE STRUCTURAL AND BIOCHEMICAL EVIDENCE FOR A BOAT-LIKE
TITLE 2 TRANSITION
TITLE 3 STATE IN BETA-MANNOSIDASES
SOURCE 2 ORGANISM_SCIENTIFIC: BACTEROIDES THETAIOTAOMICRON;
SOURCE 3 ORGANISM_TAXID: 818;
COMPND 5 EC: 3.2.1.25;
KEYWDS LINEAR FREE ENERGY RELATIONSHIP, TRANSITION STATE MIMIC,
KEYWDS 2 MANNOSIDASE, GLYCOSIDE HYDROLASE, HYDROLASE
SCOP: none
PDB: 2p3j PDBsum
HEADER VIRAL PROTEIN 09-MAR-07 2P3J
TITLE CRYSTAL STRUCTURE OF THE ARG101ALA MUTANT PROTEIN OF RHESUS
TITLE 2 ROTAVIRUS VP8*
SOURCE 2 ORGANISM_SCIENTIFIC: RHESUS ROTAVIRUS;
SOURCE 3 ORGANISM_TAXID: 10969;
KEYWDS BETA-SANDWICH, VIRAL PROTEIN
SCOP: none
PDB: 2z0z PDBsum
HEADER TRANSFERASE 07-MAY-07 2Z0Z
TITLE CRYSTAL STRUCTURE OF PUTATIVE ACETYLTRANSFERASE
SOURCE 2 ORGANISM_SCIENTIFIC: THERMUS THERMOPHILUS;
SOURCE 3 ORGANISM_TAXID: 300852;
COMPND 5 EC: 2.3.1.128;
KEYWDS ALPHA/BETA PROTEIN, TRANSFERASE, STRUCTURAL GENOMICS,
KEYWDS 2 NPPSFA, NATIONAL PROJECT ON PROTEIN STRUCTURAL AND
KEYWDS 3 FUNCTIONAL ANALYSES, RIKEN STRUCTURAL GENOMICS/PROTEOMICS
KEYWDS 4 INITIATIVE, RSGI
SCOP: none