##########
# PROTEIN: Q2246_545_165.5wLII_11172_44 384 165 # Category_B
# Length: 384
# Weight: 41645.94
# Similarity: UniProt_B6AN94, BLAST_B6AN94, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
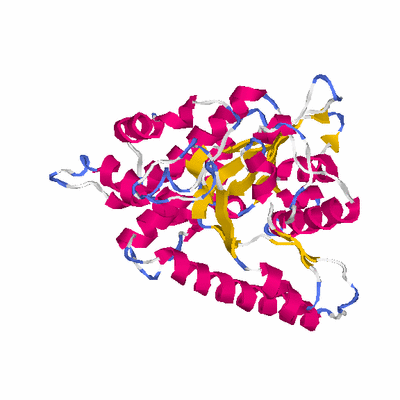
# Cat 80.99 M_00 3cqy_A 370 2 e-116 32.000 311:331 (1-331:54-364)
# Evl 80.73 M_00 3cqy_A 370 2 e-113 33.000 310:332 (1-331:54-364)
# Sid 74.48 M_00 3cqy_A 370 2 5e-47 35.000 286:309 (25-331:78-365)
# Cov 80.99 M_00 3cqy_A 370 2 e-116 32.000 311:331 (1-331:54-364)
# LaL 11.20 M_02 2pan_A 616 6 0.064 23.000 43:46 (246-288:215-260)
# OvN 80.99 M_00 3cqy_A 370 2 e-116 32.000 311:331 (1-331:54-364)
# OvC 80.99 M_00 3cqy_A 370 2 e-116 32.000 311:331 (1-331:54-364)
PDB: 3cqy PDBsum
HEADER TRANSFERASE 03-APR-08 3CQY
TITLE CRYSTAL STRUCTURE OF A FUNCTIONALLY UNKNOWN PROTEIN
TITLE 2 (SO_1313) FROM SHEWANELLA ONEIDENSIS MR-1
SOURCE 2 ORGANISM_SCIENTIFIC: SHEWANELLA ONEIDENSIS MR-1;
SOURCE 3 ORGANISM_TAXID: 211586;
COMPND 5 EC: 2.7.1.-;
KEYWDS APC7501, SO_1313, STRUCTURAL GENOMICS, PSI-2, SHEWANELLA
KEYWDS 2 ONEIDENSIS MR-1, PROTEIN STRUCTURE INITIATIVE, MIDWEST
KEYWDS 3 CENTER FOR STRUCTURAL GENOMICS, MCSG, ATP-BINDING,
KEYWDS 4 CARBOHYDRATE METABOLISM, KINASE, NUCLEOTIDE-BINDING,
KEYWDS 5 TRANSFERASE
SCOP: none
PDB: 2pan PDBsum
HEADER LYASE 27-MAR-07 2PAN
TITLE CRYSTAL STRUCTURE OF E. COLI GLYOXYLATE CARBOLIGASE
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 5 EC: 4.1.1.47;
KEYWDS THIAMIN-DIPHOSPHATE (THDP), THIMAIN-DEPENDENT ENZYMES, FAD,
KEYWDS 2 ENZYME, GLYOXYLATE CARBOLIGASE, LYASE
SCOP: none