##########
# PROTEIN: Q2246_545_174.5wLII_11172_66 133 174 # Category_C2
# Length: 133
# Weight: 15698.67
# Similarity: UniProt_B6ANB6, BLAST_B6ANB6, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 75.94 M_00 2hnh_A 910 1 8e-18 10.000 101:117 (14-127:167-270)
# Evl 75.94 M_00 2hnh_A 910 1 8e-18 10.000 101:117 (14-127:167-270)
# Sid 34.59 M_00 z02v 256 0 0.094 25.000 46:47 (1-47:1-46)
# Cov 81.95 M_04 z0n6 800 0 4e-15 6.000 109:119 (11-129:164-272)
# LaL 34.59 M_00 z02v 256 0 0.094 25.000 46:47 (1-47:1-46)
# OvN 34.59 M_00 z02v 256 0 0.094 25.000 46:47 (1-47:1-46)
# OvC 81.95 M_04 z0n6 800 0 4e-15 6.000 109:119 (11-129:164-272)
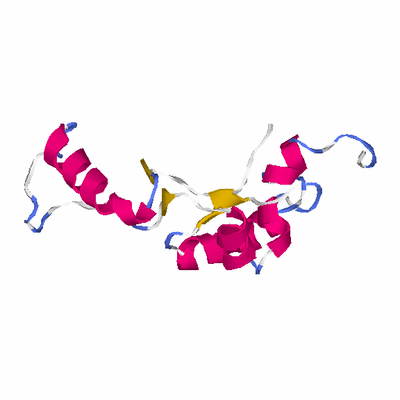
PDB: 2hnh PDBsum
HEADER TRANSFERASE 12-JUL-06 2HNH
TITLE CRYSTAL STRUCTURE OF THE CATALYTIC ALPHA SUBUNIT OF E. COLI
TITLE 2 REPLICATIVE DNA POLYMERASE III
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 5 EC: 2.7.7.7;
KEYWDS DNA POLYMERASE III, DNA REPLICATION, NUCLEOTIDYLTRANSFERASE,
KEYWDS 2 POL BETA, PHP
SCOP: none
PDB: 2ap1 PDBsum
HEADER TRANSFERASE 15-AUG-05 2AP1
TITLE CRYSTAL STRUCTURE OF THE PUTATIVE REGULATORY PROTEIN
SOURCE 2 ORGANISM_SCIENTIFIC: SALMONELLA TYPHIMURIUM;
SOURCE 3 ORGANISM_TAXID: 602;
KEYWDS ZINC BINDING PROTEIN, STRUCTURAL GENOMICS, PSI, PROTEIN
KEYWDS 2 STRUCTURE INITIATIVE, MIDWEST CENTER FOR STRUCTURAL
KEYWDS 3 GENOMICS, MCSG, PUTATIVE KINASE, TRANSFERASE
SCOP: c.55 Ribonuclease H-like motif
SCOP: c.55.1 Actin-like ATPase domain
SCOP: c.55.1.10 ROK