##########
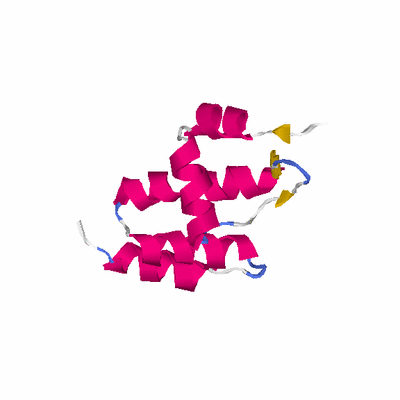
# PROTEIN: Q2246_545_176.5wLII_11172_72 108 176 # Category_C1
# Length: 108
# Weight: 12849.82
# Similarity: UniProt_B6ANC1, BLAST_B6ANC1, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 77.78 M_01 1q06_B 135 6 4e-22 18.000 84:88 (21-106:1-87)
# Evl 77.78 M_00 z0jy 132 0 1e-24 13.000 84:88 (21-106:1-87)
# Sid 77.78 M_01 1q06_B 135 6 4e-22 18.000 84:88 (21-106:1-87)
# Cov 84.26 M_03 3d6z_A 284 2 5e-17 11.000 91:104 (16-108:1-103)
# LaL 24.07 M_10 2dg6_A 222 1 9e-05 11.000 26:26 (25-50:5-30)
# OvN 54.63 M_11 z08y 744 0 0.016 9.000 59:77 (4-62:533-609)
# OvC 82.41 M_02 1r8d_A 109 3 4e-17 12.000 89:93 (19-108:1-92)
PDB: 1q06 PDBsum
HEADER TRANSCRIPTION 15-JUL-03 1Q06
TITLE CRYSTAL STRUCTURE OF THE AG(I) FORM OF E. COLI CUER, A
TITLE 2 COPPER EFFLUX REGULATOR
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS MERR FAMILY TRANSCRIPTIONAL REGULATOR, COPPER EFFLUX
KEYWDS 2 REGULATOR
SCOP: a.6 Putative DNA-binding domain
SCOP: a.6.1 Putative DNA-binding domain
SCOP: a.6.1.3 DNA-binding N-terminal domain of transcription activators
PDB: 3d6z PDBsum
HEADER TRANSCRIPTION REGULATOR/DNA 20-MAY-08 3D6Z
TITLE CRYSTAL STRUCTURE OF R275E MUTANT OF BMRR BOUND TO DNA AND
TITLE 2 RHODAMINE
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS SUBTILIS;
KEYWDS MULTIDRUG RESISTANCE, TRANSCRIPTION REGULATION, PROTEIN-DNA
KEYWDS 2 COMPLEX, ACTIVATOR, DNA-BINDING, TRANSCRIPTION
KEYWDS 3 REGULATOR/DNA COMPLEX
SCOP: none
PDB: 2dg6 PDBsum
HEADER GENE REGULATION 08-MAR-06 2DG6
TITLE CRYSTAL STRUCTURE OF THE PUTATIVE TRANSCRIPTIONAL REGULATOR
TITLE 2 SCO5550 FROM STREPTOMYCES COELICOLOR A3(2)
SOURCE 2 ORGANISM_SCIENTIFIC: STREPTOMYCES COELICOLOR A3(2);
SOURCE 3 ORGANISM_TAXID: 100226;
KEYWDS WINGED-HELIX MOTIF, MERR FAMILY, GENE REGULATION
SCOP: none
PDB: 1l1l PDBsum
HEADER OXIDOREDUCTASE 18-FEB-02 1L1L
TITLE CRYSTAL STRUCTURE OF B-12 DEPENDENT (CLASS II)
TITLE 2 RIBONUCLEOTIDE REDUCTASE
SOURCE 2 ORGANISM_SCIENTIFIC: LACTOBACILLUS LEICHMANNII;
SOURCE 3 ORGANISM_TAXID: 28039;
COMPND 4 EC: 1.17.4.2;
KEYWDS 10-STRANDED ALPHA-BETA BARREL, CENTRAL FINGER LOOP,
KEYWDS 2 OXIDOREDUCTASE
SCOP: c.7 PFL-like glycyl radical enzymes
SCOP: c.7.1 PFL-like glycyl radical enzymes
SCOP: c.7.1.4 B12-dependent (class II) ribonucleotide reductase
PDB: 1r8d PDBsum
HEADER TRANSCRIPTION/DNA 23-OCT-03 1R8D
TITLE CRYSTAL STRUCTURE OF MTAN BOUND TO DNA
SOURCE 6 ORGANISM_SCIENTIFIC: BACILLUS SUBTILIS;
SOURCE 7 ORGANISM_TAXID: 1423;
KEYWDS PROTEIN-DNA COMPLEX, TRANSCRIPTION/DNA COMPLEX
SCOP: a.6 Putative DNA-binding domain
SCOP: a.6.1 Putative DNA-binding domain
SCOP: a.6.1.3 DNA-binding N-terminal domain of transcription activators