##########
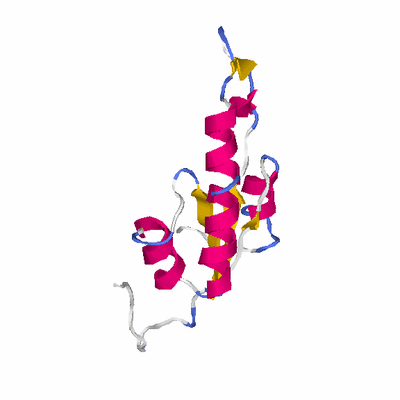
# PROTEIN: Q2246_545_27.5wLII_10776_2 183 27 # Category_C1
# Length: 183
# Weight: 21250.31
# Similarity: UniProt_B6ANE5, BLAST_B6ANE5, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 57.92 M_01 2j4d_B 525 2 3e-21 15.000 106:114 (63-171:27-137)
# Evl 57.92 M_01 2j4d_B 525 2 3e-21 15.000 106:114 (63-171:27-137)
# Sid 57.92 M_01 2j4d_B 525 2 3e-21 15.000 106:114 (63-171:27-137)
# Cov 57.92 M_01 2j4d_B 525 2 3e-21 15.000 106:114 (63-171:27-137)
# LaL 49.73 M_03 1np7_A 489 2 1e-18 10.000 91:96 (72-163:2-96)
# OvN 57.92 M_01 2j4d_B 525 2 3e-21 15.000 106:114 (63-171:27-137)
# OvC 48.63 M_06 1dnp_A 471 2 1e-16 11.000 89:104 (77-179:2-91)
PDB: 2j4d PDBsum
HEADER DNA-BINDING PROTEIN 28-AUG-06 2J4D
TITLE CRYPTOCHROME 3 FROM ARABIDOPSIS THALIANA
SOURCE 2 ORGANISM_SCIENTIFIC: ARABIDOPSIS THALIANA;
SOURCE 3 ORGANISM_COMMON: MOUSE-EAR CRESS;
SOURCE 4 ORGANISM_TAXID: 3702;
KEYWDS DNA-BINDING PROTEIN, CRYPTOCHROME, FLAVOPROTEIN, FAD,
KEYWDS 2 MITOCHONDRION, PLASTID, CHROMOPHORE, CHLOROPLAST, TRANSIT
KEYWDS 3 PEPTIDE, BLUE-LIGHT RESPONSE
SCOP: none
PDB: 1np7 PDBsum
HEADER LYASE 17-JAN-03 1NP7
TITLE CRYSTAL STRUCTURE ANALYSIS OF SYNECHOCYSTIS SP. PCC6803
TITLE 2 CRYPTOCHROME
SOURCE 2 ORGANISM_SCIENTIFIC: SYNECHOCYSTIS SP. PCC 6803;
SOURCE 3 ORGANISM_TAXID: 1148;
KEYWDS PROTEIN WITH FAD COFACTOR, LYASE
SCOP: a.99 Cryptochrome/photolyase FAD-binding domain
SCOP: a.99.1 Cryptochrome/photolyase FAD-binding domain
SCOP: a.99.1.1 Cryptochrome/photolyase FAD-binding domain
SCOP: c.28 Cryptochrome/photolyase, N-terminal domain
SCOP: c.28.1 Cryptochrome/photolyase, N-terminal domain
SCOP: c.28.1.1 Cryptochrome/photolyase, N-terminal domain
PDB: 1dnp PDBsum
HEADER LYASE (CARBON-CARBON) 03-JUL-95 1DNP
TITLE STRUCTURE OF DEOXYRIBODIPYRIMIDINE PHOTOLYASE
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562
COMPND 5 EC: 4.1.99.3;
KEYWDS DNA REPAIR, ELECTRON TRANSFER, EXCITATION ENERGY TRANSFER,
KEYWDS 2 LYASE, CARBON-CARBON, LYASE (CARBON-CARBON)
SCOP: a.99 Cryptochrome/photolyase FAD-binding domain
SCOP: a.99.1 Cryptochrome/photolyase FAD-binding domain
SCOP: a.99.1.1 Cryptochrome/photolyase FAD-binding domain
SCOP: c.28 Cryptochrome/photolyase, N-terminal domain
SCOP: c.28.1 Cryptochrome/photolyase, N-terminal domain
SCOP: c.28.1.1 Cryptochrome/photolyase, N-terminal domain