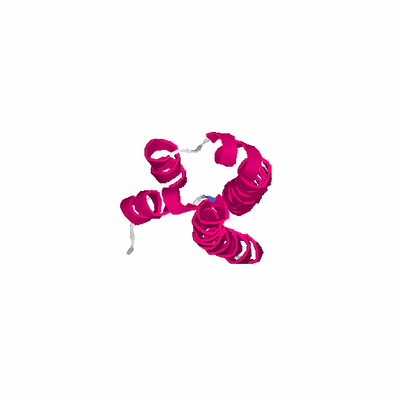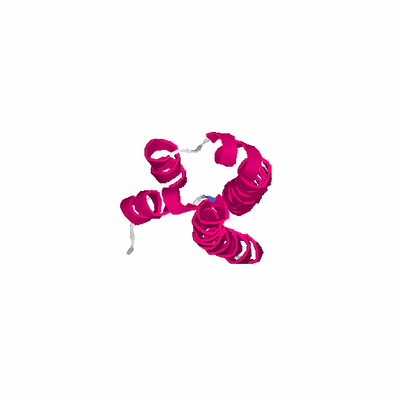##########
# PROTEIN: Q2246_545_305.5wLII_11238_83 139 305 # Category_C2
# Length: 139
# Weight: 16476.75
# Similarity: UniProt_A3ERD0, BLAST_A3ERD0, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 70.50 M_00 3epv_A 109 4 8e-08 12.000 98:98 (18-115:3-100)
# Evl 70.50 M_00 3epv_A 109 4 8e-08 12.000 98:98 (18-115:3-100)
# Sid 46.76 M_00 1knv_B 293 2 0.064 19.000 65:71 (50-120:19-83)
# Cov 82.01 M_10 1y1u_A 585 3 0.064 10.000 114:130 (20-135:19-146)
# LaL 71.22 M_00 3epv_A 109 4 5e-07 12.000 99:99 (17-115:2-100)
# OvN 56.83 M_04 2dq0_A 455 10 0.024 10.000 79:88 (9-94:18-98)
# OvC 82.01 M_10 1y1u_A 585 3 0.064 10.000 114:130 (20-135:19-146)
PDB: 3epv PDBsum
HEADER METAL BINDING PROTEIN 30-SEP-08 3EPV
TITLE X-RAY STRUCTURE OF THE METAL-SENSOR CNRX IN BOTH THE APO-
TITLE 2 AND COPPER-BOUND FORMS
SOURCE 2 ORGANISM_SCIENTIFIC: RALSTONIA METALLIDURANS CH34;
SOURCE 3 ORGANISM_TAXID: 266264;
KEYWDS ALL ALPHA HELIX, COBALT, COILED COIL, NICKEL, PERIPLASM,
KEYWDS 2 PLASMID, METAL BINDING PROTEIN
SCOP: none
PDB: 1knv PDBsum
HEADER HYDROLASE 19-DEC-01 1KNV
TITLE BSE634I RESTRICTION ENDONUCLEASE
SOURCE 2 ORGANISM_SCIENTIFIC: GEOBACILLUS STEAROTHERMOPHILUS;
SOURCE 3 ORGANISM_TAXID: 1422;
COMPND 4 EC: 3.1.21.4;
KEYWDS RESTRICTION ENDONUCLEASE, APO-ENZYME, HYDROLASE
SCOP: c.52 Restriction endonuclease-like
SCOP: c.52.1 Restriction endonuclease-like
SCOP: c.52.1.7 Cfr10I/Bse634I
PDB: 1y1u PDBsum
HEADER SIGNALING PROTEIN 19-NOV-04 1Y1U
TITLE STRUCTURE OF UNPHOSPHORYLATED STAT5A
SOURCE 2 ORGANISM_SCIENTIFIC: MUS MUSCULUS;
SOURCE 3 ORGANISM_COMMON: HOUSE MOUSE;
SOURCE 4 ORGANISM_TAXID: 10090;
KEYWDS ACTIVATOR, STAT, DNA-BINDING, SH2 DOMAIN, TRANSCRIPTION
KEYWDS 2 REGULATION, SIGNALING PROTEIN
SCOP: none
PDB: 2dq0 PDBsum
HEADER LIGASE 18-MAY-06 2DQ0
TITLE CRYSTAL STRUCTURE OF SERYL-TRNA SYNTHETASE FROM PYROCOCCUS
TITLE 2 HORIKOSHII COMPLEXED WITH A SERYL-ADENYLATE ANALOG
SOURCE 2 ORGANISM_SCIENTIFIC: PYROCOCCUS HORIKOSHII;
SOURCE 3 ORGANISM_TAXID: 70601;
COMPND 5 EC: 6.1.1.11;
KEYWDS COILED-COIL, HOMODIMER, STRUCTURAL GENOMICS, NPPSFA,
KEYWDS 2 NATIONAL PROJECT ON PROTEIN STRUCTURAL AND FUNCTIONAL
KEYWDS 3 ANALYSES, RIKEN STRUCTURAL GENOMICS/PROTEOMICS INITIATIVE,
KEYWDS 4 RSGI, LIGASE
SCOP: none