##########
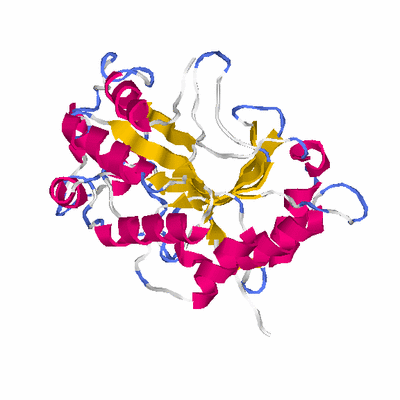
# PROTEIN: Q2246_545_405.5wLII_11277_145 311 405 # Category_C1
# Length: 311
# Weight: 35293.31
# Similarity: UniProt_B6AM01, BLAST_B6AM01, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 77.49 M_00 3c8f_A 245 2 4e-32 15.000 241:291 (14-303:2-243)
# Evl 82.64 M_00 1tv8_A 340 6 2e-63 11.000 257:304 (25-308:5-281)
# Sid 80.06 M_02 2yx0_A 342 1 2e-31 16.000 249:297 (8-298:47-309)
# Cov 89.39 M_00 1r30_B 369 2 2e-43 10.000 278:316 (1-311:35-317)
# LaL 15.43 M_11 1zjj_B 263 2 0.067 12.000 48:49 (122-169:20-68)
# OvN 89.39 M_00 1r30_B 369 2 2e-43 10.000 278:316 (1-311:35-317)
# OvC 89.39 M_00 1r30_B 369 2 2e-43 10.000 278:316 (1-311:35-317)
PDB: 3c8f PDBsum
HEADER OXIDOREDUCTASE 11-FEB-08 3C8F
TITLE 4FE-4S-PYRUVATE FORMATE-LYASE ACTIVATING ENZYME WITH
TITLE 2 PARTIALLY DISORDERED ADOMET
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
COMPND 6 EC: 1.97.1.4;
KEYWDS ADOMET RADICAL, SAM RADICAL, ACTIVASE, GLYCYL RADICAL, 4FE-
KEYWDS 2 4S, CARBOHYDRATE METABOLISM, CYTOPLASM, GLUCOSE METABOLISM,
KEYWDS 3 IRON, IRON-SULFUR, METAL-BINDING, OXIDOREDUCTASE, S-
KEYWDS 4 ADENOSYL-L-METHIONINE
SCOP: none
PDB: 1tv8 PDBsum
HEADER LIGAND BINDING PROTEIN 28-JUN-04 1TV8
TITLE STRUCTURE OF MOAA IN COMPLEX WITH S-ADENOSYLMETHIONINE
SOURCE 2 ORGANISM_SCIENTIFIC: STAPHYLOCOCCUS AUREUS;
SOURCE 3 ORGANISM_TAXID: 1280;
KEYWDS TIM BARREL, LIGAND BINDING PROTEIN
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.28 Radical SAM enzymes
SCOP: c.1.28.3 MoCo biosynthesis proteins
PDB: 2yx0 PDBsum
HEADER METAL BINDING PROTEIN 23-APR-07 2YX0
TITLE CRYSTAL STRUCTURE OF P. HORIKOSHII TYW1
SOURCE 2 ORGANISM_SCIENTIFIC: PYROCOCCUS HORIKOSHII;
SOURCE 3 ORGANISM_TAXID: 53953;
KEYWDS RADICAL SAM ENZYME, PREDICTED TRNA MODIFICATION ENZYME,
KEYWDS 2 METAL BINDING PROTEIN, STRUCTURAL GENOMICS, NPPSFA,
KEYWDS 3 NATIONAL PROJECT ON PROTEIN STRUCTURAL AND FUNCTIONAL
KEYWDS 4 ANALYSES, RIKEN STRUCTURAL GENOMICS/PROTEOMICS INITIATIVE,
KEYWDS 5 RSGI
SCOP: none
PDB: 1r30 PDBsum
HEADER TRANSFERASE 30-SEP-03 1R30
TITLE THE CRYSTAL STRUCTURE OF BIOTIN SYNTHASE, AN S-
TITLE 2 ADENOSYLMETHIONINE-DEPENDENT RADICAL ENZYME
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 5 EC: 2.8.1.6;
KEYWDS SAM RADICAL PROTEIN, TIM BARREL, FES CLUSTER, TRANSFERASE
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.28 Radical SAM enzymes
SCOP: c.1.28.1 Biotin synthase
PDB: 1zjj PDBsum
HEADER HYDROLASE 29-APR-05 1ZJJ
TITLE CRYSTAL STRUCTURE OF HYPOTHETICAL PROTEIN PH1952 FROM
TITLE 2 PYROCOCCUS HORIKOSHII OT3
SOURCE 2 ORGANISM_SCIENTIFIC: PYROCOCCUS HORIKOSHII OT3;
SOURCE 3 ORGANISM_TAXID: 70601;
KEYWDS ALPHA/BETA HYDROLASE FOLD, HAD SUPERFAMILY, STRUCTURAL
KEYWDS 2 GENOMICS, RIKEN STRUCTURAL GENOMICS/PROTEOMICS INITIATIVE,
KEYWDS 3 RSGI, NPPSFA, NATIONAL PROJECT ON PROTEIN STRUCTURAL AND
KEYWDS 4 FUNCTIONAL ANALYSES
SCOP: none