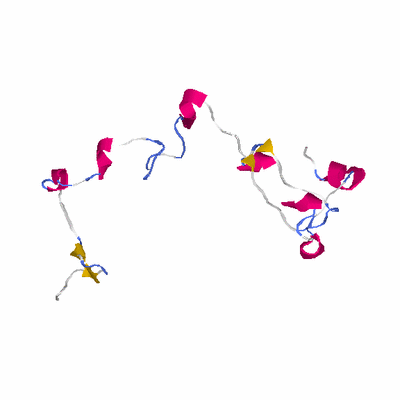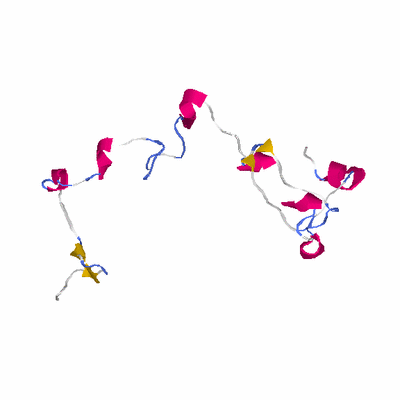##########
# PROTEIN: Q2246_545_420.5wLII_11277_201 113 420 # Category_B
# Length: 113
# Weight: 12873.92
# Similarity: UniProt_A3EPJ9, BLAST_A3EPJ9, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 88.50 M_01 5req_A 727 2 3e-28 20.000 100:118 (6-106:35-151)
# Evl 86.73 M_00 3bic_A 762 2 1e-30 17.000 98:120 (4-106:47-163)
# Sid 48.67 M_00 5req_A 727 2 0.038 35.000 55:60 (10-69:43-97)
# Cov 96.46 M_01 1rjd_C 334 10 2e-13 10.000 109:146 (1-109:74-219)
# LaL 48.67 M_00 5req_A 727 2 0.038 35.000 55:60 (10-69:43-97)
# OvN 96.46 M_01 1rjd_C 334 10 2e-13 10.000 109:146 (1-109:74-219)
# OvC 92.04 M_00 z0pa 379 0 4e-30 13.000 104:121 (2-112:70-183)
PDB: 5req PDBsum
HEADER ISOMERASE 03-AUG-98 5REQ
TITLE METHYLMALONYL-COA MUTASE, Y89F MUTANT, SUBSTRATE COMPLEX
SOURCE 2 ORGANISM_SCIENTIFIC: PROPIONIBACTERIUM FREUDENREICHII
SOURCE 4 ORGANISM_TAXID: 1752;
SOURCE 15 ORGANISM_SCIENTIFIC: PROPIONIBACTERIUM FREUDENREICHII
COMPND 4 EC: 5.4.99.2;
COMPND 9 EC: 5.4.99.2;
KEYWDS ISOMERASE, MUTASE, INTRAMOLECULAR TRANSFERASE
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.19 Cobalamin (vitamin B12)-dependent enzymes
SCOP: c.1.19.1 Methylmalonyl-CoA mutase, N-terminal (CoA-binding) domain
SCOP: c.23 Flavodoxin-like
SCOP: c.23.6 Cobalamin (vitamin B12)-binding domain
SCOP: c.23.6.1 Cobalamin (vitamin B12)-binding domain
PDB: 3bic PDBsum
HEADER ISOMERASE 30-NOV-07 3BIC
TITLE CRYSTAL STRUCTURE OF HUMAN METHYLMALONYL-COA MUTASE
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 7 EC: 5.4.99.2;
KEYWDS ORGANIC ACIDURIA, METHYLMALONYL COA MUTASE DEFICIENCY,
KEYWDS 2 METABOLIC DISEASE, STRUCTURAL GENOMICS, STRUCTURAL GENOMICS
KEYWDS 3 CONSORTIUM, SGC, COBALAMIN, COBALT, DISEASE MUTATION,
KEYWDS 4 ISOMERASE, METAL-BINDING, MITOCHONDRION, POLYMORPHISM,
KEYWDS 5 TRANSIT PEPTIDE
SCOP: none
PDB: 1rjd PDBsum
HEADER TRANSFERASE 19-NOV-03 1RJD
TITLE STRUCTURE OF PPM1, A LEUCINE CARBOXY METHYLTRANSFERASE
TITLE 2 INVOLVED IN THE REGULATION OF PROTEIN PHOSPHATASE 2A
TITLE 3 ACTIVITY
SOURCE 2 ORGANISM_SCIENTIFIC: SACCHAROMYCES CEREVISIAE;
SOURCE 3 ORGANISM_COMMON: BAKER'S YEAST;
SOURCE 4 ORGANISM_TAXID: 4932;
COMPND 6 EC: 2.1.1.-;
KEYWDS SAM DEPENDENT METHYLTRANSFERASE
SCOP: c.66 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1 S-adenosyl-L-methionine-dependent methyltransferases
SCOP: c.66.1.37 Leucine carboxy methyltransferase Ppm1
PDB: 1jdb PDBsum
HEADER LIGASE 25-MAR-97 1JDB
TITLE CARBAMOYL PHOSPHATE SYNTHETASE FROM ESCHERICHIA COLI
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
SOURCE 5 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
COMPND 4 EC: 6.3.5.5;
COMPND 8 EC: 6.3.5.5
KEYWDS LIGASE, AMIDOTRANSFERASE, SYNTHASE
SCOP: a.92 Carbamoyl phosphate synthetase, large subunit connection domain
SCOP: a.92.1 Carbamoyl phosphate synthetase, large subunit connection domain
SCOP: a.92.1.1 Carbamoyl phosphate synthetase, large subunit connection domain
SCOP: c.8 The "swivelling" beta/beta/alpha domain
SCOP: c.8.3 Carbamoyl phosphate synthetase, small subunit N-terminal domain
SCOP: c.8.3.1 Carbamoyl phosphate synthetase, small subunit N-terminal domain
SCOP: c.23 Flavodoxin-like
SCOP: c.23.16 Class I glutamine amidotransferase-like
SCOP: c.23.16.1 Class I glutamine amidotransferases (GAT)
SCOP: c.24 Methylglyoxal synthase-like
SCOP: c.24.1 Methylglyoxal synthase-like
SCOP: c.24.1.1 Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain
SCOP: c.30 PreATP-grasp domain
SCOP: c.30.1 PreATP-grasp domain
SCOP: c.30.1.1 BC N-terminal domain-like
SCOP: d.142 ATP-grasp
SCOP: d.142.1 Glutathione synthetase ATP-binding domain-like
SCOP: d.142.1.2 BC ATP-binding domain-like