##########
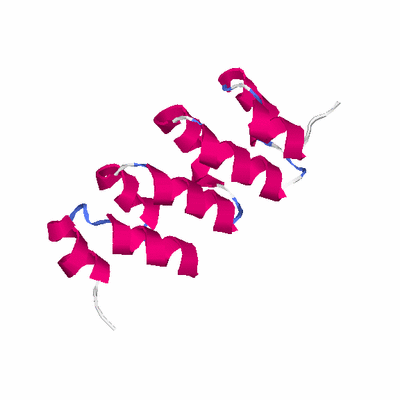
# PROTEIN: Q2246_545_437.5wLII_11277_265 132 437 # Category_C1
# Length: 132
# Weight: 15194.85
# Similarity: UniProt_B6AMC2, BLAST_B6AMC2, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 67.42 M_05 1lrv_A 244 1 0.012 15.000 89:93 (38-130:91-179)
# Evl 96.97 M_00 2yvx_A 473 4 1e-23 12.000 128:131 (3-130:32-162)
# Sid 37.88 M_05 2vf7_A 842 5 0.023 19.000 50:52 (4-53:408-459)
# Cov 98.48 M_00 2yvx_A 473 4 1e-19 14.000 130:135 (2-132:30-163)
# LaL 51.52 M_05 2a6m_B 155 4 0.039 10.000 68:68 (36-103:25-92)
# OvN 97.73 M_00 2yvx_A 473 4 3e-23 12.000 129:132 (2-130:31-162)
# OvC 98.48 M_00 2yvx_A 473 4 1e-19 14.000 130:135 (2-132:30-163)
PDB: 1lrv PDBsum
HEADER LEUCINE-RICH REPEATS 05-NOV-96 1LRV
TITLE A LEUCINE-RICH REPEAT VARIANT WITH A NOVEL REPETITIVE
TITLE 2 PROTEIN STRUCTURAL MOTIF
SOURCE 2 ORGANISM_SCIENTIFIC: AZOTOBACTER VINELANDII;
SOURCE 3 ORGANISM_TAXID: 354;
KEYWDS LEUCINE-RICH REPEATS, REPETITIVE STRUCTURE, IRON SULFUR
KEYWDS 2 PROTEINS, NITROGEN FIXATION
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.1 ARM repeat
SCOP: a.118.1.5 Leucine-rich repeat variant
PDB: 2yvx PDBsum
HEADER TRANSPORT PROTEIN 18-APR-07 2YVX
TITLE CRYSTAL STRUCTURE OF MAGNESIUM TRANSPORTER MGTE
SOURCE 2 ORGANISM_SCIENTIFIC: THERMUS THERMOPHILUS HB8;
SOURCE 3 ORGANISM_TAXID: 300852;
KEYWDS MEMBRANE PROTEIN, TRANSPORT PROTEIN
SCOP: none
PDB: 2vf7 PDBsum
HEADER DNA-BINDING PROTEIN 31-OCT-07 2VF7
TITLE CRYSTAL STRUCTURE OF UVRA2 FROM DEINOCOCCUS RADIODURANS
SOURCE 2 ORGANISM_SCIENTIFIC: DEINOCOCCUS RADIODURANS;
SOURCE 3 ORGANISM_TAXID: 1299;
KEYWDS DNA-BINDING PROTEIN, NUCLEOTIDE-BINDING, ZINC-BINDING
KEYWDS 2 DOMAIN, SOS RESPONSE, METAL-BINDING, EXCISION NUCLEASE,
KEYWDS 3 ZINC-FINGER, ATP-BINDING, DNA-BINDING, DNA EXCISION, ZINC,
KEYWDS 4 CYTOPLASM, DNA DAMAGE, DNA REPAIR, ABC PROTEIN DNA-BINDING
KEYWDS 5 PROTEIN
SCOP: none
PDB: 2a6m PDBsum
HEADER TRANSCRIPTION/DNA 03-JUL-05 2A6M
TITLE CRYSTAL STRUCTURE OF THE ISHP608 TRANSPOSASE
SOURCE 2 ORGANISM_SCIENTIFIC: HELICOBACTER PYLORI;
SOURCE 3 ORGANISM_TAXID: 210;
KEYWDS RNA RECOGNITION MOTIF, TRANSCRIPTION/DNA COMPLEX
SCOP: d.58 Ferredoxin-like
SCOP: d.58.57 Transposase IS200-like
SCOP: d.58.57.1 Transposase IS200-like