##########
# PROTEIN: Q2246_545_550.5wLII_11391_14 218 550 # Category_B
# Length: 218
# Weight: 23956.48
# Similarity: UniProt_B6AS31, BLAST_B6AS31, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 75.23 M_00 1y0g_A 191 4 7e-63 35.000 164:192 (31-217:6-191)
# Evl 75.23 M_00 1y0g_A 191 4 7e-63 35.000 164:192 (31-217:6-191)
# Sid 74.31 M_00 1y0g_A 191 4 7e-25 38.000 162:174 (47-215:22-189)
# Cov 75.23 M_00 1y0g_A 191 4 7e-63 35.000 164:192 (31-217:6-191)
# LaL 40.37 M_04 3cb9_A 248 2 0.028 13.000 88:91 (109-197:85-174)
# OvN 75.23 M_00 1y0g_A 191 4 3e-52 35.000 164:194 (29-217:4-191)
# OvC 75.23 M_00 1y0g_A 191 4 7e-63 35.000 164:192 (31-217:6-191)
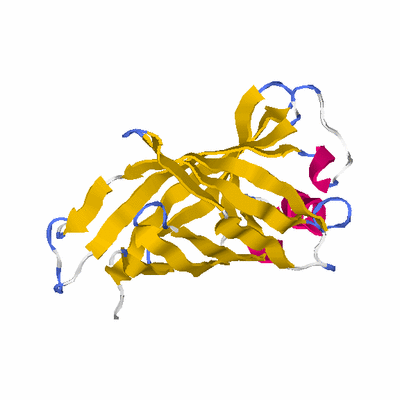
PDB: 1y0g PDBsum
HEADER LIPID BINDING PROTEIN 15-NOV-04 1Y0G
TITLE CRYSTAL STRUCTURE OF THE ESCHERICHIA COLI YCEI PROTEIN,
TITLE 2 STRUCTURAL GENOMICS
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS LIPID BINDING PROTEIN, LIPOCALIN, COFACTOR, COENZYME,
KEYWDS 2 DEHYDROGENASE, HYDROLASE, PREDICTED, STRUCTURAL GENOMICS,
KEYWDS 3 PROTEIN STRUCTURE INITIATIVE, PSI, NYSGXRC, NEW YORK SGX
KEYWDS 4 RESEARCH CENTER FOR STRUCTURAL GENOMICS
SCOP: b.61 Streptavidin-like
SCOP: b.61.6 YceI-like
SCOP: b.61.6.1 YceI-like
PDB: 3cb9 PDBsum
HEADER LUMINESCENT PROTEIN 21-FEB-08 3CB9
TITLE DEVELOPMENT OF A FAMILY OF REDOX-SENSITIVE GREEN
TITLE 2 FLUORESCENT PROTEIN INDICATORS FOR USE IN RELATIVELY
TITLE 3 OXIDIZING SUBCELLULAR ENVIRONMENTS
SOURCE 2 ORGANISM_SCIENTIFIC: AEQUOREA VICTORIA;
SOURCE 3 ORGANISM_COMMON: JELLYFISH;
SOURCE 4 ORGANISM_TAXID: 6100;
KEYWDS GFP, REDOX, LUMINESCENT PROTEIN
SCOP: none