##########
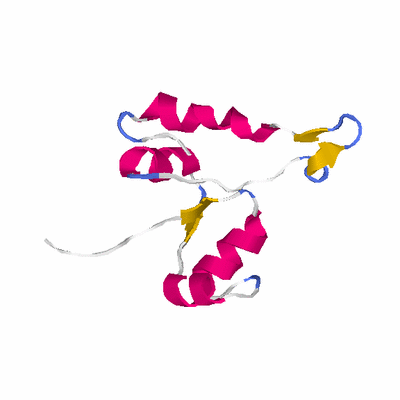
# PROTEIN: Q2246_545_60.5wLII_10961_3 123 60 # Category_C1
# Length: 123
# Weight: 13162.90
# Similarity: UniProt_A3ES63, BLAST_A3ES63, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 65.85 M_06 1dli_A 402 1 7e-08 18.000 81:93 (22-105:22-111)
# Evl 75.61 M_00 2o3j_A 481 3 5e-25 13.000 93:112 (1-98:18-124)
# Sid 65.85 M_06 1dli_A 402 1 7e-08 18.000 81:93 (22-105:22-111)
# Cov 78.86 M_04 3g79_A 478 2 2e-14 13.000 97:119 (1-102:27-140)
# LaL 22.76 M_10 1zoy_B 252 2 0.059 14.000 28:28 (78-105:108-135)
# OvN 75.61 M_00 2o3j_A 481 3 5e-25 13.000 93:112 (1-98:18-124)
# OvC 76.42 M_09 1ad3_A 452 2 0.008 14.000 94:114 (19-113:335-447)
PDB: 1dli PDBsum
HEADER OXIDOREDUCTASE 09-DEC-99 1DLI
TITLE THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH)
TITLE 2 REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD
TITLE 3 OXIDATION
SOURCE 2 ORGANISM_SCIENTIFIC: STREPTOCOCCUS PYOGENES;
SOURCE 3 ORGANISM_TAXID: 1314;
COMPND 4 EC: 1.1.1.22;
KEYWDS ROSSMANN FOLD, TERNARY COMPLEX, CRYSTALLOGRAPHIC DIMER,
KEYWDS 2 OXIDOREDUCTASE
SCOP: a.100 6-phosphogluconate dehydrogenase C-terminal domain-like
SCOP: a.100.1 6-phosphogluconate dehydrogenase C-terminal domain-like
SCOP: a.100.1.4 UDP-glucose/GDP-mannose dehydrogenase dimerisation domain
SCOP: c.2 NAD(P)-binding Rossmann-fold domains
SCOP: c.2.1 NAD(P)-binding Rossmann-fold domains
SCOP: c.2.1.6 6-phosphogluconate dehydrogenase-like, N-terminal domain
SCOP: c.26 Adenine nucleotide alpha hydrolase-like
SCOP: c.26.3 UDP-glucose/GDP-mannose dehydrogenase C-terminal domain
SCOP: c.26.3.1 UDP-glucose/GDP-mannose dehydrogenase C-terminal domain
PDB: 2o3j PDBsum
HEADER OXIDOREDUCTASE 01-DEC-06 2O3J
TITLE STRUCTURE OF CAENORHABDITIS ELEGANS UDP-GLUCOSE
TITLE 2 DEHYDROGENASE
SOURCE 2 ORGANISM_SCIENTIFIC: CAENORHABDITIS ELEGANS;
SOURCE 3 ORGANISM_TAXID: 6239;
COMPND 6 EC: 1.1.1.22;
KEYWDS UDP-GLUCOSE DEHYDROGENASE, STRUCTURAL GENOMICS, PSI-2,
KEYWDS 2 PROTEIN STRUCTURE INITIATIVE, NEW YORK SGX RESEARCH CENTER
KEYWDS 3 FOR STRUCTURAL GENOMICS, NYSGXRC, OXIDOREDUCTASE
SCOP: none
PDB: 3g79 PDBsum
HEADER OXIDOREDUCTASE 09-FEB-09 3G79
TITLE CRYSTAL STRUCTURE OF NDP-N-ACETYL-D-GALACTOSAMINURONIC ACID
TITLE 2 DEHYDROGENASE FROM METHANOSARCINA MAZEI GO1
SOURCE 2 ORGANISM_SCIENTIFIC: METHANOSARCINA MAZEI GO1;
SOURCE 3 ORGANISM_TAXID: 192952;
COMPND 6 EC: 1.1.1.-;
KEYWDS STRUCTURAL GENOMICS, CRYSTAL STRUCTURE, PROTEIN STRUCTURE
KEYWDS 2 INITIATIVE, NEW YORK STRUCTURAL GENOMIX RESEARCH CONSORTIUM,
KEYWDS 3 NYSGXRC, NDP-N-ACETYL-D-GALACTOSAMINURONIC ACID
KEYWDS 4 DEHYDROGENASE, OXIDOREDUCTASE, PSI-2, NEW YORK SGX RESEARCH
KEYWDS 5 CENTER FOR STRUCTURAL GENOMICS
SCOP: none
PDB: 1zoy PDBsum
HEADER OXIDOREDUCTASE 15-MAY-05 1ZOY
TITLE CRYSTAL STRUCTURE OF MITOCHONDRIAL RESPIRATORY COMPLEX II
TITLE 2 FROM PORCINE HEART AT 2.4 ANGSTROMS
SOURCE 2 ORGANISM_SCIENTIFIC: SUS SCROFA;
SOURCE 3 ORGANISM_COMMON: PIG;
SOURCE 4 ORGANISM_TAXID: 9823;
COMPND 5 EC: 1.3.5.1;
COMPND 9 EC: 1.3.5.1;
KEYWDS SUCCINATE, UBIQUINONE OXIDOREDUCTASE, MITOCHONDRIAL
KEYWDS 2 RESPIRATORY COMPLEX II, MEMBRANE PROTEIN STRUCTURE
SCOP: none
PDB: 1ad3 PDBsum
HEADER OXIDOREDUCTASE 25-JUN-96 1AD3
TITLE CLASS 3 ALDEHYDE DEHYDROGENASE COMPLEX WITH NICOTINAMIDE-
TITLE 2 ADENINE-DINUCLEOTIDE
SOURCE 2 ORGANISM_SCIENTIFIC: RATTUS NORVEGICUS;
SOURCE 3 ORGANISM_COMMON: NORWAY RAT;
SOURCE 4 ORGANISM_TAXID: 10116;
COMPND 5 EC: 1.2.1.5;
KEYWDS NADP, OXIDOREDUCTASE, AROMATIC ALDEHYDE
SCOP: c.82 ALDH-like
SCOP: c.82.1 ALDH-like
SCOP: c.82.1.1 ALDH-like