##########
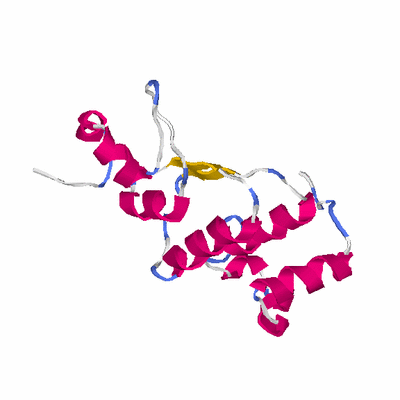
# PROTEIN: Q2246_545_72.5wLII_10961_40 191 72 # Category_C1
# Length: 191
# Weight: 21476.77
# Similarity: UniProt_B6AQA5, BLAST_B6AQA5, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 65.45 M_04 2ix3_A 986 4 7e-24 15.000 125:132 (10-137:519-647)
# Evl 52.88 M_00 z0b6 242 0 3e-25 10.000 101:108 (22-126:121-224)
# Sid 19.90 M_01 1gte_D 1025 16 0.005 23.000 38:38 (57-94:346-383)
# Cov 65.45 M_04 2ix3_A 986 4 7e-24 15.000 125:132 (10-137:519-647)
# LaL 19.90 M_01 1gte_D 1025 16 0.005 23.000 38:38 (57-94:346-383)
# OvN 61.78 M_16 2it1_A 362 2 3e-14 11.000 118:135 (1-129:103-226)
# OvC 65.45 M_04 2ix3_A 986 4 7e-24 15.000 125:132 (10-137:519-647)
PDB: 2ix3 PDBsum
HEADER TRANSLATION 06-JUL-06 2IX3
TITLE STRUCTURE OF YEAST ELONGATION FACTOR 3
SOURCE 2 ORGANISM_SCIENTIFIC: SACCHAROMYCES CEREVISIAE;
SOURCE 3 ORGANISM_COMMON: BAKER'S YEAST;
SOURCE 4 ORGANISM_TAXID: 4932;
KEYWDS PROTEIN BIOSYNTHESIS, TRANSLATION, ELONGATION FACTOR,
KEYWDS 2 PHOSPHORYLATION, NUCLEOTIDE-BINDING, RRNA-BINDING, RNA-
KEYWDS 3 BINDING, ACETYLATION, ATP-BINDING
SCOP: none
PDB: 1b0u PDBsum
HEADER TRANSPORT PROTEIN 12-NOV-98 1B0U
TITLE ATP-BINDING SUBUNIT OF THE HISTIDINE PERMEASE FROM
TITLE 2 SALMONELLA TYPHIMURIUM
SOURCE 2 ORGANISM_SCIENTIFIC: SALMONELLA TYPHIMURIUM;
SOURCE 3 ORGANISM_TAXID: 602;
KEYWDS ABC TRANSPORTER, HISTIDINE PERMEASE, TRANSPORT PROTEIN
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.12 ABC transporter ATPase domain-like
PDB: 1gte PDBsum
HEADER OXIDOREDUCTASE 15-JAN-02 1GTE
TITLE DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, BINARY
TITLE 2 COMPLEX WITH 5-IODOURACIL
SOURCE 2 ORGANISM_SCIENTIFIC: SUS SCROFA;
SOURCE 3 ORGANISM_COMMON: PIG;
SOURCE 4 ORGANISM_TAXID: 9823;
COMPND 6 EC: 1.3.1.2;
KEYWDS ELECTRON TRANSFER, FLAVIN, IRON-SULFUR CLUSTERS, PYRIMIDINE
KEYWDS 2 CATABOLISM, 5-FLUOROURACIL DEGRADATION, OXIDOREDUCTASE
SCOP: a.1 Globin-like
SCOP: a.1.2 alpha-helical ferredoxin
SCOP: a.1.2.2 Dihydropyrimidine dehydrogenase, N-terminal domain
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.4 FMN-linked oxidoreductases
SCOP: c.1.4.1 FMN-linked oxidoreductases
SCOP: c.3 FAD/NAD(P)-binding domain
SCOP: c.3.1 FAD/NAD(P)-binding domain
SCOP: c.3.1.1 C-terminal domain of adrenodoxin reductase-like
SCOP: c.4 Nucleotide-binding domain
SCOP: c.4.1 Nucleotide-binding domain
SCOP: c.4.1.1 N-terminal domain of adrenodoxin reductase-like
SCOP: d.58 Ferredoxin-like
SCOP: d.58.1 4Fe-4S ferredoxins
SCOP: d.58.1.5 Ferredoxin domains from multidomain proteins
PDB: 2it1 PDBsum
HEADER TRANSPORT PROTEIN 18-OCT-06 2IT1
TITLE STRUCTURE OF PH0203 PROTEIN FROM PYROCOCCUS HORIKOSHII
SOURCE 2 ORGANISM_SCIENTIFIC: PYROCOCCUS HORIKOSHII;
SOURCE 3 ORGANISM_TAXID: 53953;
KEYWDS ATP BINDING PROTEIN, STRUCTURAL GENOMICS, NPPSFA, NATIONAL
KEYWDS 2 PROJECT ON PROTEIN STRUCTURAL AND FUNCTIONAL ANALYSES,
KEYWDS 3 RIKEN STRUCTURAL GENOMICS/PROTEOMICS INITIATIVE, RSGI,
KEYWDS 4 TRANSPORT PROTEIN
SCOP: none