LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_129.5wLII_11077_43
Total number of 3D structures: 36
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
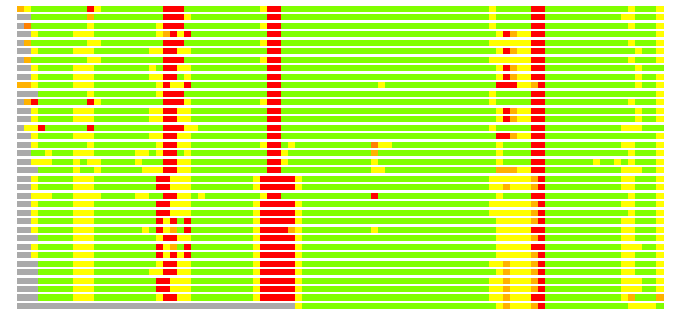
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 2j1g_E |
218 |
93 |
85 |
1.22 |
15.29 |
88.988 |
6.429 |
T P |
| 2j61_A |
216 |
93 |
84 |
1.16 |
15.48 |
87.658 |
6.674 |
T P |
| 2j5z_A |
214 |
93 |
85 |
1.31 |
16.47 |
87.601 |
6.020 |
T P |
| 1re3_E |
301 |
93 |
84 |
1.51 |
10.71 |
87.347 |
5.225 |
T P |
| 2jhk_F |
217 |
93 |
85 |
1.40 |
14.12 |
87.314 |
5.668 |
T P |
| 3e1i_B |
298 |
93 |
84 |
1.46 |
11.90 |
87.276 |
5.394 |
T P |
| 2jhm_F |
217 |
93 |
85 |
1.40 |
14.12 |
87.234 |
5.657 |
T P |
| 1m1j_B |
402 |
93 |
84 |
1.48 |
10.71 |
87.215 |
5.330 |
T P |
| 3bvh_B |
298 |
93 |
84 |
1.47 |
11.90 |
87.215 |
5.336 |
T P |
| 2oyh_B |
298 |
93 |
85 |
1.66 |
10.59 |
87.116 |
4.840 |
T P |
| 2d39_C |
212 |
93 |
83 |
0.94 |
16.87 |
87.012 |
7.988 |
T P |
| 2j3o_B |
218 |
93 |
83 |
1.13 |
15.66 |
86.969 |
6.775 |
T P |
| 2z4e_B |
303 |
93 |
84 |
1.46 |
11.90 |
86.932 |
5.394 |
T P |
| 1fzc_B |
308 |
93 |
84 |
1.49 |
11.90 |
86.776 |
5.272 |
T P |
| 2j3f_D |
217 |
93 |
83 |
1.18 |
14.46 |
86.521 |
6.485 |
T P |
| 1deq_B |
380 |
93 |
83 |
1.48 |
12.05 |
86.347 |
5.240 |
T P |
| 1jc9_A |
220 |
93 |
85 |
1.56 |
12.94 |
86.031 |
5.125 |
T P |
| 1z3s_A |
216 |
93 |
85 |
1.58 |
12.94 |
84.923 |
5.063 |
T P |
| 2gy7_A |
216 |
93 |
85 |
1.46 |
12.94 |
84.861 |
5.463 |
T P |
| 1lwu_B |
315 |
93 |
84 |
1.71 |
9.52 |
84.522 |
4.642 |
T P |
| 2fib_A |
250 |
93 |
83 |
1.64 |
9.64 |
84.102 |
4.759 |
T P |
| 1n73_C |
322 |
93 |
83 |
1.66 |
12.05 |
84.036 |
4.719 |
T P |
| 1z3u_A |
216 |
93 |
84 |
1.50 |
13.10 |
83.979 |
5.263 |
T P |
| 3fib_A |
249 |
93 |
83 |
1.66 |
9.64 |
83.942 |
4.718 |
T P |
| 1lwu_C |
317 |
93 |
83 |
1.66 |
12.05 |
83.909 |
4.708 |
T P |
| 1rf1_C |
298 |
93 |
82 |
1.66 |
9.76 |
83.465 |
4.663 |
T P |
| 1deq_C |
370 |
93 |
83 |
1.80 |
9.64 |
83.418 |
4.361 |
T P |
| 3bvh_C |
293 |
93 |
82 |
1.64 |
9.76 |
83.329 |
4.708 |
T P |
| 2hlo_C |
292 |
93 |
82 |
1.71 |
9.76 |
83.310 |
4.522 |
T P |
| 2oyh_C |
299 |
93 |
82 |
1.65 |
9.76 |
83.234 |
4.680 |
T P |
| 1m1j_C |
390 |
93 |
82 |
1.67 |
10.98 |
83.206 |
4.632 |
T P |
| 2h43_C |
290 |
93 |
82 |
1.69 |
9.76 |
82.980 |
4.593 |
T P |
| 1fzc_C |
301 |
93 |
82 |
1.69 |
9.76 |
82.819 |
4.574 |
T P |
| 1ei3_C |
397 |
93 |
82 |
1.69 |
10.98 |
82.819 |
4.574 |
T P |
| 1re3_F |
291 |
93 |
81 |
1.68 |
9.88 |
82.739 |
4.557 |
T P |
| 1fzd_A |
197 |
93 |
52 |
1.73 |
11.54 |
52.009 |
2.844 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

