LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_152.5wLII_11111_93
Total number of 3D structures: 41
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
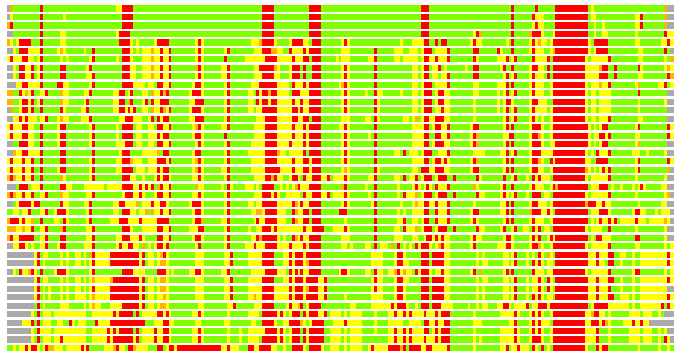
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 2qc5_A |
298 |
227 |
195 |
0.70 |
24.62 |
85.223 |
24.400 |
T P |
| 2z2n_A |
293 |
227 |
194 |
0.99 |
22.68 |
83.608 |
17.819 |
T P |
| 2z2p_A |
293 |
227 |
194 |
1.03 |
22.16 |
83.103 |
17.241 |
T P |
| 2z2o_C |
299 |
227 |
192 |
1.17 |
21.35 |
82.618 |
15.160 |
T P |
| 1l0q_A |
391 |
227 |
175 |
1.77 |
21.14 |
68.663 |
9.378 |
T P |
| 3dm0_A |
675 |
227 |
177 |
1.93 |
6.21 |
60.544 |
8.712 |
T P |
| 1vyh_C |
310 |
227 |
183 |
2.05 |
5.46 |
60.063 |
8.515 |
T P |
| 2h9l_A |
321 |
227 |
172 |
1.91 |
5.23 |
58.878 |
8.578 |
T P |
| 2gnq_A |
316 |
227 |
173 |
1.91 |
5.78 |
58.434 |
8.605 |
T P |
| 3frx_B |
313 |
227 |
174 |
2.04 |
6.32 |
57.745 |
8.121 |
T P |
| 2bcj_B |
339 |
227 |
178 |
2.06 |
5.62 |
57.402 |
8.240 |
T P |
| 1tbg_A |
340 |
227 |
175 |
2.00 |
6.29 |
57.290 |
8.320 |
T P |
| 1got_B |
339 |
227 |
176 |
2.01 |
6.82 |
57.103 |
8.338 |
T P |
| 2pbi_D |
354 |
227 |
176 |
2.01 |
7.39 |
56.755 |
8.358 |
T P |
| 2h9m_A |
304 |
227 |
171 |
1.90 |
7.60 |
56.541 |
8.533 |
T P |
| 2h6n_B |
305 |
227 |
172 |
1.93 |
6.98 |
56.484 |
8.454 |
T P |
| 2co0_A |
304 |
227 |
172 |
1.91 |
7.56 |
56.415 |
8.571 |
T P |
| 1erj_C |
357 |
227 |
175 |
1.98 |
9.71 |
56.168 |
8.411 |
T P |
| 2cnx_A |
306 |
227 |
172 |
1.99 |
7.56 |
55.840 |
8.248 |
T P |
| 2g9a_A |
310 |
227 |
172 |
1.98 |
7.56 |
55.369 |
8.274 |
T P |
| 2g99_A |
304 |
227 |
174 |
2.03 |
6.90 |
54.580 |
8.179 |
T P |
| 1nr0_A |
610 |
227 |
173 |
2.07 |
3.47 |
54.458 |
7.956 |
T P |
| 3emh_A |
300 |
227 |
175 |
2.09 |
8.00 |
54.106 |
8.008 |
T P |
| 1a0r_B |
339 |
227 |
170 |
2.00 |
6.47 |
53.821 |
8.100 |
T P |
| 2ovr_B |
442 |
227 |
170 |
2.11 |
8.82 |
53.477 |
7.685 |
T P |
| 1nex_B |
444 |
227 |
171 |
2.15 |
4.68 |
53.296 |
7.596 |
T P |
| 2h14_A |
303 |
227 |
176 |
2.09 |
6.25 |
52.938 |
8.029 |
T P |
| 2hes_X |
308 |
227 |
167 |
1.99 |
7.78 |
52.893 |
7.998 |
T P |
| 3fm0_A |
328 |
227 |
176 |
2.13 |
7.39 |
52.811 |
7.896 |
T P |
| 2iaq_A |
312 |
227 |
164 |
2.18 |
11.59 |
52.727 |
7.204 |
T P |
| 2iau_A |
312 |
227 |
165 |
2.20 |
10.91 |
51.822 |
7.187 |
T P |
| 1p22_A |
402 |
227 |
170 |
2.11 |
6.47 |
51.696 |
7.677 |
T P |
| 2iax_A |
312 |
227 |
165 |
2.23 |
12.12 |
51.605 |
7.084 |
T P |
| 2iao_A |
312 |
227 |
164 |
2.24 |
10.37 |
51.097 |
7.006 |
T P |
| 2iap_A |
312 |
227 |
165 |
2.27 |
11.52 |
50.673 |
6.972 |
T P |
| 2iav_A |
312 |
227 |
163 |
2.29 |
13.50 |
49.906 |
6.818 |
T P |
| 2dg0_A |
322 |
227 |
166 |
2.39 |
7.83 |
49.422 |
6.678 |
T P |
| 1pjx_A |
314 |
227 |
158 |
2.23 |
9.49 |
48.146 |
6.781 |
T P |
| 2dg1_B |
322 |
227 |
166 |
2.39 |
9.64 |
48.005 |
6.655 |
T P |
| 2dso_C |
323 |
227 |
163 |
2.36 |
9.82 |
47.916 |
6.633 |
T P |
| 2zkq_a |
306 |
227 |
153 |
2.43 |
8.50 |
44.846 |
6.055 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

