LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_156.5wLII_11111_110
Total number of 3D structures: 84
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
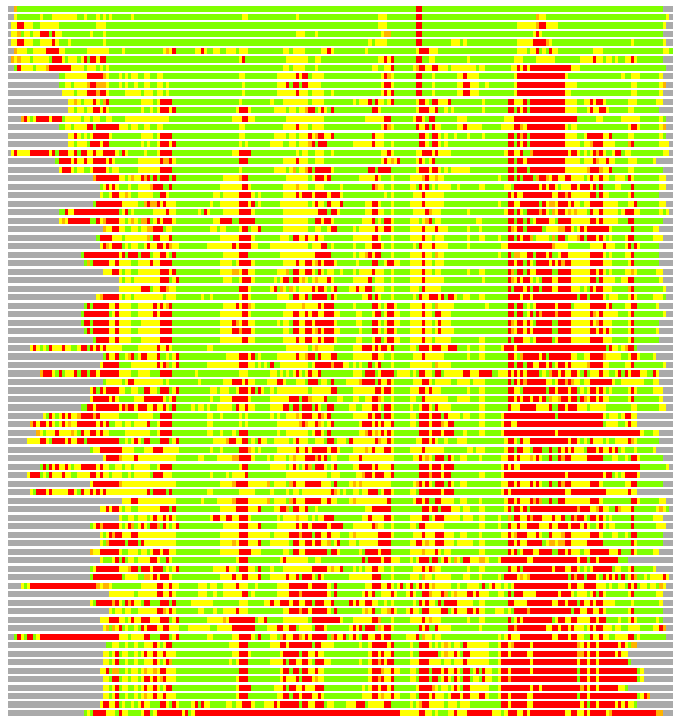
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 2avd_A |
219 |
210 |
203 |
0.48 |
23.65 |
96.230 |
34.737 |
T P |
| 2hnk_B |
232 |
210 |
205 |
1.46 |
20.00 |
90.988 |
13.131 |
T P |
| 3c3y_A |
225 |
210 |
201 |
1.41 |
19.90 |
90.351 |
13.328 |
T P |
| 3cbg_A |
218 |
210 |
199 |
1.36 |
19.10 |
90.115 |
13.671 |
T P |
| 1sui_A |
227 |
210 |
198 |
1.49 |
21.21 |
87.859 |
12.489 |
T P |
| 3dr5_A |
216 |
210 |
196 |
1.88 |
17.35 |
81.812 |
9.909 |
T P |
| 3duw_A |
220 |
210 |
191 |
1.83 |
18.32 |
81.732 |
9.886 |
T P |
| 3c3p_A |
198 |
210 |
178 |
1.83 |
19.10 |
76.293 |
9.201 |
T P |
| 2cl5_A |
214 |
210 |
164 |
1.86 |
15.85 |
68.854 |
8.383 |
T P |
| 3bwm_A |
214 |
210 |
164 |
1.88 |
12.80 |
68.849 |
8.282 |
T P |
| 3bwy_A |
214 |
210 |
163 |
1.90 |
11.66 |
68.248 |
8.163 |
T P |
| 1o54_A |
265 |
210 |
157 |
2.00 |
13.38 |
64.630 |
7.492 |
T P |
| 2yvl_A |
247 |
210 |
149 |
1.88 |
14.77 |
62.864 |
7.536 |
T P |
| 2gpy_B |
192 |
210 |
168 |
2.10 |
21.43 |
62.606 |
7.630 |
T P |
| 2zlb_A |
212 |
210 |
161 |
2.01 |
15.53 |
60.490 |
7.641 |
T P |
| 2pwy_A |
251 |
210 |
149 |
1.95 |
12.75 |
59.528 |
7.257 |
T P |
| 1i9g_A |
264 |
210 |
153 |
1.94 |
16.34 |
59.249 |
7.502 |
T P |
| 2as0_A |
396 |
210 |
155 |
1.94 |
11.61 |
58.598 |
7.580 |
T P |
| 1l3i_A |
186 |
210 |
151 |
2.06 |
15.89 |
58.101 |
6.976 |
T P |
| 2b25_A |
254 |
210 |
143 |
1.97 |
12.59 |
53.848 |
6.892 |
T P |
| 3g5l_A |
222 |
210 |
144 |
2.17 |
6.94 |
52.412 |
6.338 |
T P |
| 3e7p_A |
253 |
210 |
149 |
2.16 |
13.42 |
50.884 |
6.606 |
T P |
| 3dli_A |
221 |
210 |
139 |
2.01 |
9.35 |
49.047 |
6.581 |
T P |
| 1wzn_A |
244 |
210 |
142 |
2.25 |
14.79 |
48.926 |
6.042 |
T P |
| 2p8j_B |
207 |
210 |
148 |
2.24 |
10.14 |
48.824 |
6.319 |
T P |
| 3dtn_A |
220 |
210 |
139 |
2.23 |
11.51 |
48.181 |
5.964 |
T P |
| 1y8c_A |
246 |
210 |
142 |
2.26 |
14.08 |
47.858 |
6.015 |
T P |
| 2o57_A |
282 |
210 |
147 |
2.32 |
12.24 |
47.819 |
6.073 |
T P |
| 2yqz_A |
261 |
210 |
140 |
2.27 |
11.43 |
47.176 |
5.901 |
T P |
| 3bkw_A |
219 |
210 |
136 |
2.13 |
15.44 |
47.080 |
6.105 |
T P |
| 1kph_B |
285 |
210 |
137 |
2.12 |
9.49 |
46.812 |
6.159 |
T P |
| 3bus_A |
251 |
210 |
146 |
2.29 |
13.70 |
46.635 |
6.114 |
T P |
| 1xxl_A |
234 |
210 |
142 |
2.27 |
9.15 |
46.545 |
5.989 |
T P |
| 1vl5_A |
231 |
210 |
138 |
2.14 |
9.42 |
46.444 |
6.163 |
T P |
| 1p91_A |
268 |
210 |
128 |
2.06 |
14.06 |
46.382 |
5.924 |
T P |
| 1kpi_A |
291 |
210 |
138 |
2.27 |
7.25 |
46.015 |
5.817 |
T P |
| 2fk8_A |
281 |
210 |
137 |
2.18 |
8.76 |
45.745 |
6.008 |
T P |
| 1kpg_A |
285 |
210 |
136 |
2.09 |
8.82 |
45.651 |
6.204 |
T P |
| 1l1e_A |
272 |
210 |
133 |
2.13 |
9.02 |
45.447 |
5.971 |
T P |
| 1tpy_A |
285 |
210 |
138 |
2.18 |
10.87 |
45.353 |
6.046 |
T P |
| 1i1n_A |
224 |
210 |
132 |
2.29 |
15.15 |
45.240 |
5.512 |
T P |
| 1ve3_B |
226 |
210 |
134 |
2.25 |
11.19 |
44.834 |
5.711 |
T P |
| 3e23_A |
198 |
210 |
140 |
2.26 |
10.71 |
44.791 |
5.941 |
T P |
| 3bkx_A |
274 |
210 |
137 |
2.41 |
9.49 |
44.681 |
5.458 |
T P |
| 3dp7_A |
351 |
210 |
137 |
2.40 |
11.68 |
44.665 |
5.471 |
T P |
| 3d2l_C |
242 |
210 |
135 |
2.25 |
11.11 |
44.663 |
5.755 |
T P |
| 3f4k_A |
254 |
210 |
137 |
2.27 |
12.41 |
44.607 |
5.781 |
T P |
| 2gh1_A |
281 |
210 |
136 |
2.27 |
11.76 |
44.546 |
5.743 |
T P |
| 1dl5_A |
317 |
210 |
127 |
2.28 |
16.54 |
44.441 |
5.340 |
T P |
| 2yxe_A |
214 |
210 |
127 |
2.23 |
15.75 |
44.355 |
5.441 |
T P |
| 1vbf_B |
226 |
210 |
128 |
2.33 |
14.06 |
44.129 |
5.270 |
T P |
| 1sqg_A |
424 |
210 |
141 |
2.27 |
16.31 |
44.121 |
5.942 |
T P |
| 3bxo_A |
236 |
210 |
136 |
2.28 |
16.18 |
44.031 |
5.706 |
T P |
| 1d2g_A |
292 |
210 |
142 |
2.52 |
15.49 |
44.030 |
5.412 |
T P |
| 1jg1_A |
215 |
210 |
122 |
2.05 |
10.66 |
43.834 |
5.685 |
T P |
| 1r18_A |
223 |
210 |
132 |
2.36 |
14.39 |
43.474 |
5.372 |
T P |
| 1xva_A |
292 |
210 |
142 |
2.51 |
16.20 |
43.242 |
5.447 |
T P |
| 2pbf_A |
218 |
210 |
130 |
2.33 |
16.92 |
43.204 |
5.347 |
T P |
| 1r8x_B |
284 |
210 |
140 |
2.48 |
17.14 |
42.870 |
5.417 |
T P |
| 1nkv_C |
249 |
210 |
125 |
2.18 |
12.80 |
42.737 |
5.476 |
T P |
| 1ri5_A |
252 |
210 |
136 |
2.37 |
11.03 |
42.600 |
5.511 |
T P |
| 2avn_A |
247 |
210 |
135 |
2.34 |
9.63 |
42.192 |
5.541 |
T P |
| 2azt_B |
277 |
210 |
140 |
2.53 |
15.71 |
42.156 |
5.318 |
T P |
| 1r74_B |
279 |
210 |
133 |
2.44 |
18.05 |
41.892 |
5.233 |
T P |
| 3e8s_A |
220 |
210 |
127 |
2.24 |
14.17 |
41.805 |
5.433 |
T P |
| 1g6q_1 |
328 |
210 |
111 |
2.12 |
11.71 |
41.789 |
5.002 |
T P |
| 2gs9_A |
211 |
210 |
125 |
2.15 |
13.60 |
41.532 |
5.545 |
T P |
| 3dlc_A |
219 |
210 |
129 |
2.39 |
13.95 |
41.091 |
5.190 |
T P |
| 1adm_A |
415 |
210 |
132 |
2.47 |
12.88 |
41.002 |
5.145 |
T P |
| 1aqj_B |
383 |
210 |
131 |
2.39 |
17.56 |
40.603 |
5.267 |
T P |
| 3dh0_B |
190 |
210 |
125 |
2.23 |
9.60 |
40.559 |
5.369 |
T P |
| 2ih2_A |
393 |
210 |
126 |
2.30 |
18.25 |
40.219 |
5.254 |
T P |
| 1vlm_A |
207 |
210 |
126 |
2.26 |
10.32 |
40.051 |
5.330 |
T P |
| 1qzz_A |
340 |
210 |
127 |
2.40 |
14.96 |
40.045 |
5.072 |
T P |
| 3ccf_B |
242 |
210 |
128 |
2.30 |
10.94 |
40.023 |
5.344 |
T P |
| 3b3f_A |
337 |
210 |
110 |
2.32 |
13.64 |
38.555 |
4.542 |
T P |
| 1or8_A |
313 |
210 |
108 |
2.23 |
15.74 |
38.375 |
4.630 |
T P |
| 2v74_D |
332 |
210 |
108 |
2.21 |
12.96 |
38.096 |
4.685 |
T P |
| 2fyt_A |
311 |
210 |
105 |
2.17 |
11.43 |
37.662 |
4.619 |
T P |
| 1f3l_A |
321 |
210 |
104 |
2.17 |
13.46 |
37.647 |
4.575 |
T P |
| 1orh_A |
318 |
210 |
106 |
2.21 |
16.04 |
37.061 |
4.593 |
T P |
| 1ori_A |
316 |
210 |
106 |
2.23 |
11.32 |
36.955 |
4.546 |
T P |
| 3b3j_A |
332 |
210 |
107 |
2.38 |
9.35 |
34.800 |
4.315 |
T P |
| 3dnx_A |
146 |
210 |
54 |
2.78 |
9.26 |
16.342 |
1.872 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

