LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_169.5wLII_11172_58
Total number of 3D structures: 59
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
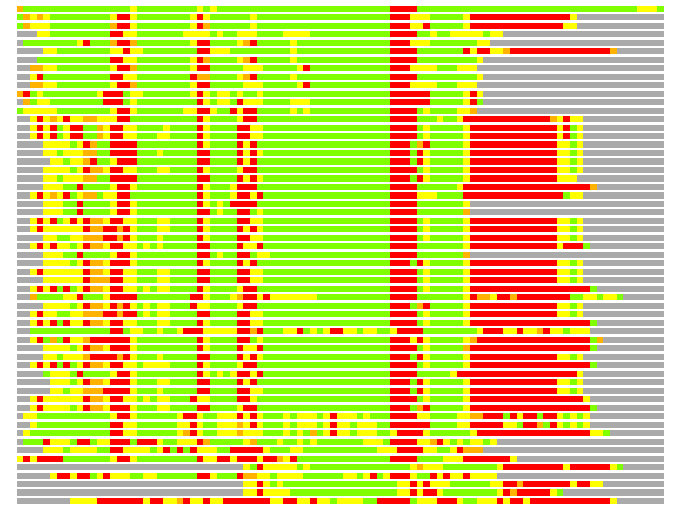
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 3bk7_A |
593 |
97 |
93 |
1.01 |
23.66 |
93.403 |
8.397 |
T P |
| 2c3m_A |
1231 |
97 |
63 |
1.81 |
25.40 |
60.243 |
3.298 |
T P |
| 2c42_A |
1231 |
97 |
63 |
1.85 |
25.40 |
60.030 |
3.229 |
T P |
| 3c8y_A |
574 |
97 |
64 |
1.97 |
23.44 |
58.207 |
3.086 |
T P |
| 1jb0_C |
80 |
97 |
61 |
1.82 |
16.39 |
57.574 |
3.183 |
T P |
| 1jnr_B |
149 |
97 |
61 |
1.87 |
27.87 |
57.484 |
3.098 |
T P |
| 2vkr_A |
103 |
97 |
59 |
1.66 |
25.42 |
56.990 |
3.351 |
T P |
| 1h7x_B |
1019 |
97 |
59 |
1.85 |
25.42 |
56.634 |
3.029 |
T P |
| 1xer_A |
102 |
97 |
59 |
1.74 |
27.12 |
56.624 |
3.200 |
T P |
| 1gte_D |
1014 |
97 |
59 |
1.85 |
25.42 |
56.507 |
3.032 |
T P |
| 3c7b_B |
363 |
97 |
58 |
1.84 |
17.24 |
55.024 |
2.984 |
T P |
| 2v4j_B |
380 |
97 |
57 |
1.87 |
22.81 |
53.327 |
2.897 |
T P |
| 1hfe_L |
396 |
97 |
58 |
2.01 |
32.76 |
52.588 |
2.753 |
T P |
| 1bqx_A |
77 |
97 |
58 |
1.98 |
24.14 |
52.393 |
2.788 |
T P |
| 7fd1_A |
106 |
97 |
56 |
1.83 |
23.21 |
52.348 |
2.900 |
T P |
| 1b0t_A |
106 |
97 |
56 |
1.83 |
23.21 |
52.339 |
2.895 |
T P |
| 1fdd_A |
106 |
97 |
56 |
1.88 |
21.43 |
52.249 |
2.825 |
T P |
| 1frk_A |
106 |
97 |
56 |
1.89 |
21.43 |
52.062 |
2.811 |
T P |
| 1fri_A |
106 |
97 |
55 |
1.81 |
21.82 |
51.605 |
2.874 |
T P |
| 1pc5_A |
106 |
97 |
58 |
2.03 |
20.69 |
51.487 |
2.720 |
T P |
| 1b0v_A |
106 |
97 |
55 |
1.87 |
21.82 |
51.433 |
2.791 |
T P |
| 1dur_A |
55 |
97 |
53 |
1.55 |
24.53 |
51.356 |
3.210 |
T P |
| 1bc6_A |
77 |
97 |
56 |
1.90 |
21.43 |
50.921 |
2.796 |
T P |
| 1fdx |
54 |
97 |
52 |
1.51 |
25.00 |
50.678 |
3.237 |
T P |
| 1fca_A |
55 |
97 |
53 |
1.57 |
22.64 |
50.525 |
3.168 |
T P |
| 1g3o_A |
106 |
97 |
57 |
2.00 |
21.05 |
50.518 |
2.710 |
T P |
| 1f5c_A |
106 |
97 |
58 |
2.05 |
20.69 |
50.505 |
2.697 |
T P |
| 1frj_A |
106 |
97 |
58 |
2.07 |
22.41 |
50.488 |
2.676 |
T P |
| 2v2k_A |
104 |
97 |
55 |
1.91 |
23.64 |
50.416 |
2.741 |
T P |
| 2fdn_A |
55 |
97 |
53 |
1.60 |
22.64 |
50.398 |
3.110 |
T P |
| 1a6l_A |
106 |
97 |
57 |
1.99 |
21.05 |
50.321 |
2.721 |
T P |
| 1d3w_A |
106 |
97 |
58 |
2.04 |
20.69 |
50.247 |
2.709 |
T P |
| 1ff2_A |
106 |
97 |
57 |
2.03 |
19.30 |
49.894 |
2.672 |
T P |
| 1pc4_A |
106 |
97 |
54 |
1.86 |
20.37 |
49.860 |
2.760 |
T P |
| 2fug_9 |
154 |
97 |
64 |
2.18 |
23.44 |
49.807 |
2.809 |
T P |
| 1fd2_A |
106 |
97 |
58 |
2.20 |
18.97 |
49.672 |
2.524 |
T P |
| 1f5b_A |
106 |
97 |
57 |
2.04 |
21.05 |
49.599 |
2.662 |
T P |
| 1gao_A |
106 |
97 |
54 |
1.88 |
20.37 |
49.586 |
2.723 |
T P |
| 1ti6_B |
274 |
97 |
64 |
2.24 |
12.50 |
49.444 |
2.731 |
T P |
| 1frh_A |
106 |
97 |
54 |
1.96 |
22.22 |
49.359 |
2.617 |
T P |
| 1h98_A |
77 |
97 |
54 |
1.90 |
24.07 |
49.341 |
2.705 |
T P |
| 1frl_A |
106 |
97 |
55 |
1.93 |
21.82 |
49.331 |
2.705 |
T P |
| 1frm_A |
106 |
97 |
54 |
1.88 |
20.37 |
49.329 |
2.728 |
T P |
| 1clf_A |
55 |
97 |
52 |
1.63 |
23.08 |
49.294 |
3.004 |
T P |
| 1ftc_A |
106 |
97 |
55 |
1.98 |
21.82 |
49.098 |
2.640 |
T P |
| 1g6b_A |
106 |
97 |
55 |
2.00 |
23.64 |
49.048 |
2.618 |
T P |
| 1frx_A |
106 |
97 |
56 |
2.07 |
19.64 |
48.319 |
2.585 |
T P |
| 2fd2_A |
106 |
97 |
54 |
1.99 |
22.22 |
48.289 |
2.589 |
T P |
| 1q16_B |
509 |
97 |
66 |
2.24 |
9.09 |
48.104 |
2.817 |
T P |
| 1y4z_B |
509 |
97 |
65 |
2.21 |
7.69 |
47.535 |
2.816 |
T P |
| 2ivf_B |
337 |
97 |
63 |
2.29 |
17.46 |
45.947 |
2.639 |
T P |
| 2o01_C |
80 |
97 |
56 |
2.17 |
14.29 |
42.737 |
2.472 |
T P |
| 1e08_A |
371 |
97 |
51 |
2.36 |
31.37 |
40.129 |
2.070 |
T P |
| 3cf4_A |
766 |
97 |
48 |
2.06 |
31.25 |
38.638 |
2.222 |
T P |
| 2vpz_B |
193 |
97 |
41 |
2.04 |
17.07 |
36.554 |
1.916 |
T P |
| 1k0t_A |
80 |
97 |
52 |
2.34 |
13.46 |
36.442 |
2.128 |
T P |
| 1kqf_B |
289 |
97 |
40 |
2.03 |
17.50 |
35.080 |
1.880 |
T P |
| 1h0h_B |
214 |
97 |
38 |
1.97 |
26.32 |
34.309 |
1.835 |
T P |
| 1yqt_A |
515 |
97 |
38 |
2.84 |
7.89 |
25.187 |
1.294 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

