LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_207.5wLII_11184_31
Total number of 3D structures: 55
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
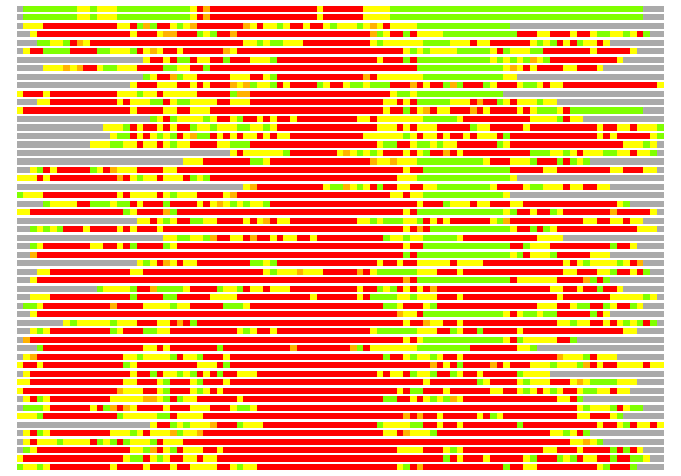
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 1hlr_A |
907 |
97 |
70 |
1.48 |
24.29 |
69.466 |
4.423 |
T P |
| 1vlb_A |
907 |
97 |
70 |
1.48 |
24.29 |
69.466 |
4.423 |
T P |
| 2dqb_C |
364 |
97 |
46 |
2.53 |
4.35 |
32.967 |
1.750 |
T P |
| 2hek_A |
369 |
97 |
39 |
2.55 |
2.56 |
29.686 |
1.471 |
T P |
| 2r6o_A |
258 |
97 |
42 |
2.56 |
9.52 |
28.888 |
1.581 |
T P |
| 1a2o_A |
347 |
97 |
39 |
2.42 |
2.56 |
28.388 |
1.548 |
T P |
| 1w25_A |
454 |
97 |
34 |
2.16 |
0.00 |
27.839 |
1.506 |
T P |
| 2gwr_A |
225 |
97 |
37 |
2.42 |
13.51 |
27.379 |
1.471 |
T P |
| 2vuh_B |
135 |
97 |
33 |
2.26 |
9.09 |
27.119 |
1.401 |
T P |
| 1ny5_B |
385 |
97 |
37 |
2.76 |
8.11 |
26.408 |
1.293 |
T P |
| 2jb9_B |
122 |
97 |
33 |
2.47 |
3.03 |
26.288 |
1.286 |
T P |
| 2oqr_A |
226 |
97 |
38 |
2.87 |
7.89 |
25.294 |
1.278 |
T P |
| 1dc7_A |
124 |
97 |
32 |
2.49 |
6.25 |
25.256 |
1.237 |
T P |
| 2hwv_A |
101 |
97 |
36 |
2.80 |
2.78 |
25.131 |
1.242 |
T P |
| 2pq7_A |
174 |
97 |
38 |
2.81 |
7.89 |
25.123 |
1.305 |
T P |
| 2pjq_B |
212 |
97 |
39 |
2.93 |
12.82 |
24.884 |
1.287 |
T P |
| 3ccg_A |
189 |
97 |
35 |
2.60 |
5.71 |
24.810 |
1.299 |
T P |
| 1kgs_A |
219 |
97 |
37 |
2.74 |
8.11 |
24.809 |
1.301 |
T P |
| 2z9m_A |
100 |
97 |
32 |
2.41 |
6.25 |
24.628 |
1.274 |
T P |
| 1ys7_B |
226 |
97 |
31 |
2.22 |
9.68 |
24.627 |
1.336 |
T P |
| 1zgz_A |
121 |
97 |
33 |
2.23 |
3.03 |
24.609 |
1.417 |
T P |
| 1s8n_A |
190 |
97 |
33 |
2.54 |
15.15 |
24.543 |
1.251 |
T P |
| 2iyn_B |
124 |
97 |
33 |
2.46 |
6.06 |
24.264 |
1.287 |
T P |
| 1yoy_A |
146 |
97 |
34 |
2.53 |
8.82 |
23.957 |
1.293 |
T P |
| 2qxy_A |
119 |
97 |
27 |
2.07 |
14.81 |
23.929 |
1.245 |
T P |
| 3b57_A |
180 |
97 |
35 |
2.68 |
8.57 |
23.820 |
1.259 |
T P |
| 1xhe_B |
122 |
97 |
30 |
2.44 |
3.33 |
23.585 |
1.182 |
T P |
| 2d1v_A |
103 |
97 |
31 |
2.72 |
3.23 |
23.407 |
1.099 |
T P |
| 3cfy_A |
130 |
97 |
28 |
2.15 |
14.29 |
23.316 |
1.247 |
T P |
| 1dc8_A |
123 |
97 |
25 |
1.90 |
4.00 |
23.238 |
1.251 |
T P |
| 2ogi_B |
194 |
97 |
32 |
2.84 |
0.00 |
22.604 |
1.090 |
T P |
| 1yio_A |
198 |
97 |
32 |
2.67 |
6.25 |
22.590 |
1.156 |
T P |
| 1zes_A |
121 |
97 |
25 |
2.14 |
16.00 |
22.431 |
1.116 |
T P |
| 2o6i_A |
436 |
97 |
33 |
2.76 |
3.03 |
22.411 |
1.154 |
T P |
| 3bre_B |
328 |
97 |
31 |
2.56 |
12.90 |
22.215 |
1.164 |
T P |
| 2j8e_A |
128 |
97 |
30 |
2.52 |
3.33 |
22.213 |
1.147 |
T P |
| 1u0s_Y |
118 |
97 |
25 |
2.22 |
16.00 |
22.045 |
1.077 |
T P |
| 2o08_A |
187 |
97 |
31 |
2.79 |
3.23 |
21.765 |
1.072 |
T P |
| 2jk1_A |
138 |
97 |
29 |
2.64 |
6.90 |
21.143 |
1.058 |
T P |
| 1dz3_A |
123 |
97 |
23 |
2.05 |
8.70 |
20.792 |
1.071 |
T P |
| 2rjn_A |
135 |
97 |
26 |
2.43 |
11.54 |
20.751 |
1.027 |
T P |
| 1b00_A |
122 |
97 |
30 |
2.78 |
0.00 |
20.490 |
1.041 |
T P |
| 1p2f_A |
217 |
97 |
29 |
2.87 |
6.90 |
20.453 |
0.977 |
T P |
| 1tmy_A |
118 |
97 |
28 |
2.59 |
3.57 |
20.266 |
1.042 |
T P |
| 2a9r_A |
117 |
97 |
27 |
2.80 |
11.11 |
20.229 |
0.931 |
T P |
| 1xhf_B |
122 |
97 |
30 |
2.85 |
16.67 |
20.070 |
1.018 |
T P |
| 1mvo_A |
121 |
97 |
28 |
2.67 |
3.57 |
19.790 |
1.011 |
T P |
| 1xx7_A |
172 |
97 |
26 |
2.63 |
7.69 |
19.582 |
0.952 |
T P |
| 2jba_B |
121 |
97 |
26 |
2.88 |
7.69 |
18.894 |
0.872 |
T P |
| 3dto_A |
189 |
97 |
28 |
2.74 |
3.57 |
18.536 |
0.984 |
T P |
| 1zh2_A |
120 |
97 |
28 |
2.95 |
3.57 |
18.380 |
0.918 |
T P |
| 2qzj_A |
121 |
97 |
25 |
2.66 |
12.00 |
17.773 |
0.906 |
T P |
| 2a9o_A |
117 |
97 |
25 |
2.73 |
8.00 |
17.649 |
0.884 |
T P |
| 2jba_A |
125 |
97 |
25 |
2.55 |
4.00 |
17.600 |
0.944 |
T P |
| 1wlf_A |
164 |
97 |
25 |
2.65 |
8.00 |
17.309 |
0.909 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

