LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_229.5wLII_11195_24
Total number of 3D structures: 33
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
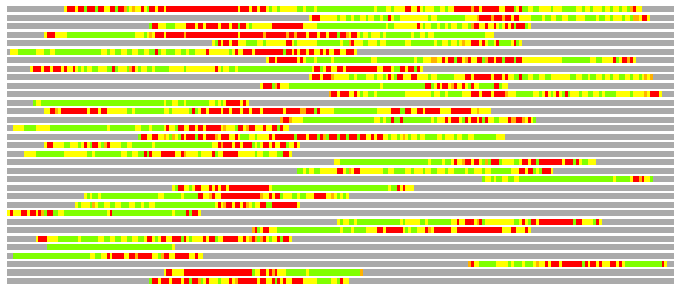
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 1c1g_A |
284 |
234 |
130 |
2.37 |
3.08 |
41.112 |
5.271 |
T P |
| 2b9c_B |
142 |
234 |
103 |
2.39 |
4.85 |
32.323 |
4.143 |
T P |
| 2v71_A |
160 |
234 |
95 |
2.48 |
3.16 |
32.134 |
3.689 |
T P |
| 2efr_A |
155 |
234 |
96 |
2.24 |
14.58 |
31.689 |
4.095 |
T P |
| 1deq_A |
180 |
234 |
95 |
2.25 |
5.26 |
31.672 |
4.035 |
T P |
| 1cii_A |
602 |
234 |
92 |
2.22 |
6.52 |
31.239 |
3.960 |
T P |
| 1hci_A |
475 |
234 |
99 |
2.24 |
5.05 |
31.050 |
4.230 |
T P |
| 1ei3_C |
397 |
234 |
100 |
2.34 |
8.00 |
30.227 |
4.099 |
T P |
| 2fxo_A |
129 |
234 |
97 |
2.51 |
9.28 |
29.796 |
3.721 |
T P |
| 3cwg_B |
507 |
234 |
73 |
1.86 |
1.37 |
28.685 |
3.734 |
T P |
| 2fxm_A |
126 |
234 |
95 |
2.41 |
5.26 |
28.623 |
3.792 |
T P |
| 1f5n_A |
570 |
234 |
70 |
1.51 |
5.71 |
28.334 |
4.351 |
T P |
| 1m1j_C |
390 |
234 |
89 |
2.55 |
5.62 |
28.246 |
3.355 |
T P |
| 1bg1_A |
559 |
234 |
73 |
2.18 |
6.85 |
26.692 |
3.202 |
T P |
| 1y1u_A |
544 |
234 |
77 |
2.15 |
9.09 |
25.840 |
3.424 |
T P |
| 2d3e_A |
130 |
234 |
84 |
2.59 |
13.10 |
25.388 |
3.127 |
T P |
| 2b5u_A |
470 |
234 |
69 |
2.28 |
8.70 |
24.416 |
2.899 |
T P |
| 2iw5_A |
666 |
234 |
78 |
2.39 |
8.97 |
24.353 |
3.130 |
T P |
| 2v1d_A |
666 |
234 |
68 |
2.06 |
11.76 |
24.013 |
3.144 |
T P |
| 1quu_A |
248 |
234 |
79 |
2.37 |
10.13 |
23.656 |
3.200 |
T P |
| 1mqm_B |
172 |
234 |
57 |
1.54 |
7.02 |
23.356 |
3.465 |
T P |
| 1ha0_A |
494 |
234 |
61 |
1.89 |
3.28 |
22.613 |
3.068 |
T P |
| 2h94_A |
647 |
234 |
69 |
2.20 |
8.70 |
22.592 |
3.000 |
T P |
| 1jch_A |
468 |
234 |
62 |
2.19 |
9.68 |
22.591 |
2.707 |
T P |
| 1qvr_A |
803 |
234 |
56 |
1.68 |
7.14 |
22.486 |
3.138 |
T P |
| 2hko_A |
647 |
234 |
70 |
2.32 |
11.43 |
22.431 |
2.892 |
T P |
| 2z3y_A |
643 |
234 |
61 |
2.20 |
9.84 |
21.756 |
2.651 |
T P |
| 2dw4_A |
634 |
234 |
65 |
2.52 |
9.23 |
20.799 |
2.484 |
T P |
| 1h6k_C |
733 |
234 |
45 |
1.17 |
8.89 |
18.504 |
3.552 |
T P |
| 1ses_A |
421 |
234 |
47 |
1.99 |
10.64 |
17.054 |
2.244 |
T P |
| 2dq3_A |
425 |
234 |
50 |
2.31 |
2.00 |
16.078 |
2.074 |
T P |
| 1lrz_A |
400 |
234 |
40 |
2.10 |
7.50 |
14.017 |
1.820 |
T P |
| 2wb7_A |
525 |
234 |
39 |
2.77 |
10.26 |
11.359 |
1.360 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

