LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_248.5wLII_11212_72
Total number of 3D structures: 104
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
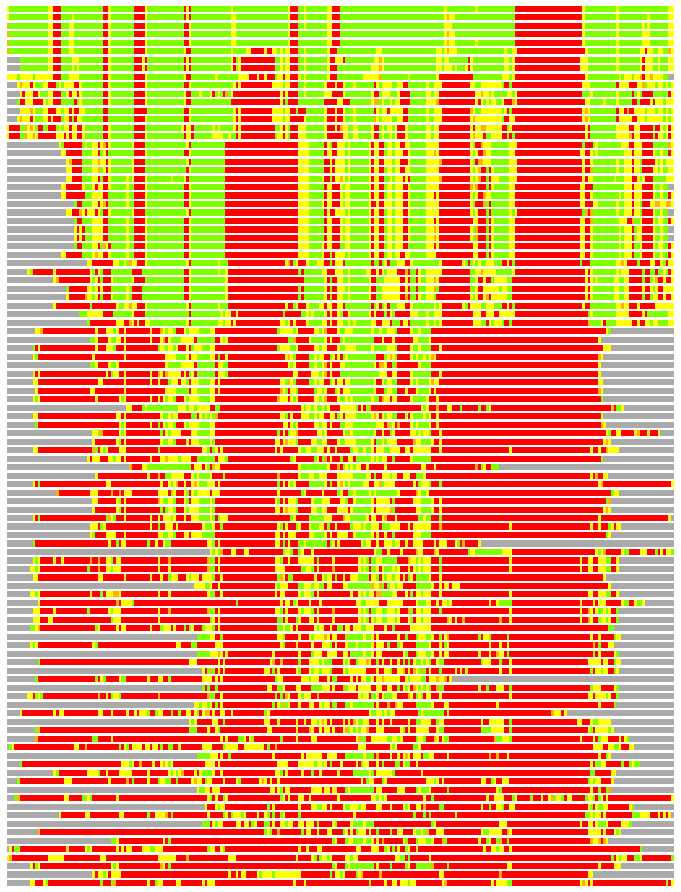
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 1m53_A |
556 |
256 |
213 |
0.99 |
23.00 |
80.863 |
19.547 |
T P |
| 2pwf_C |
556 |
256 |
213 |
1.20 |
19.25 |
79.679 |
16.349 |
T P |
| 2pwe_A |
556 |
256 |
213 |
1.20 |
19.72 |
79.622 |
16.364 |
T P |
| 1zja_A |
557 |
256 |
213 |
1.23 |
19.72 |
79.506 |
15.978 |
T P |
| 2pwh_A |
556 |
256 |
213 |
1.23 |
19.72 |
79.485 |
16.014 |
T P |
| 1uok_A |
558 |
256 |
205 |
1.51 |
12.68 |
74.979 |
12.756 |
T P |
| 2zic_A |
536 |
256 |
193 |
1.58 |
12.44 |
70.319 |
11.465 |
T P |
| 2zid_A |
536 |
256 |
193 |
1.57 |
12.44 |
70.223 |
11.570 |
T P |
| 2ze0_A |
531 |
256 |
187 |
1.88 |
12.83 |
63.827 |
9.443 |
T P |
| 1g5a_A |
628 |
256 |
170 |
2.13 |
8.24 |
50.949 |
7.633 |
T P |
| 1wza_A |
488 |
256 |
155 |
1.85 |
11.61 |
47.459 |
7.935 |
T P |
| 1lwj_A |
441 |
256 |
147 |
1.82 |
10.20 |
47.327 |
7.646 |
T P |
| 1jgi_A |
628 |
256 |
166 |
2.10 |
8.43 |
46.521 |
7.549 |
T P |
| 1mvy_A |
628 |
256 |
170 |
2.17 |
7.65 |
46.277 |
7.491 |
T P |
| 3czl_A |
612 |
256 |
155 |
2.10 |
9.68 |
44.678 |
7.048 |
T P |
| 3czg_A |
607 |
256 |
152 |
2.14 |
9.87 |
43.418 |
6.792 |
T P |
| 1gvi_A |
588 |
256 |
138 |
2.01 |
9.42 |
42.993 |
6.532 |
T P |
| 1vfo_A |
585 |
256 |
138 |
1.99 |
8.70 |
42.974 |
6.596 |
T P |
| 1j0h_A |
588 |
256 |
141 |
2.08 |
9.22 |
42.797 |
6.463 |
T P |
| 1sma_A |
588 |
256 |
140 |
2.04 |
9.29 |
42.399 |
6.546 |
T P |
| 1ea9_C |
583 |
256 |
140 |
2.07 |
11.43 |
41.992 |
6.466 |
T P |
| 1g1y_A |
585 |
256 |
132 |
1.96 |
10.61 |
41.117 |
6.421 |
T P |
| 1jf5_A |
585 |
256 |
135 |
2.01 |
10.37 |
41.002 |
6.410 |
T P |
| 2d2o_A |
585 |
256 |
134 |
2.08 |
9.70 |
40.967 |
6.137 |
T P |
| 1j0j_A |
588 |
256 |
136 |
2.03 |
9.56 |
40.896 |
6.390 |
T P |
| 1wzl_A |
585 |
256 |
131 |
1.95 |
10.69 |
40.732 |
6.391 |
T P |
| 1ji2_A |
585 |
256 |
133 |
2.00 |
10.53 |
40.384 |
6.328 |
T P |
| 1wzk_A |
585 |
256 |
133 |
2.01 |
10.53 |
40.345 |
6.300 |
T P |
| 1wzm_A |
585 |
256 |
132 |
1.93 |
11.36 |
40.320 |
6.513 |
T P |
| 1jf6_A |
585 |
256 |
134 |
1.98 |
10.45 |
40.230 |
6.443 |
T P |
| 1eha_A |
557 |
256 |
127 |
2.01 |
8.66 |
38.044 |
6.025 |
T P |
| 1cgw_A |
686 |
256 |
130 |
2.14 |
11.54 |
37.902 |
5.798 |
T P |
| 1cxl_A |
686 |
256 |
122 |
1.96 |
13.11 |
37.775 |
5.930 |
T P |
| 1cyg_A |
680 |
256 |
127 |
2.07 |
12.60 |
37.615 |
5.864 |
T P |
| 1uks_A |
686 |
256 |
124 |
2.08 |
12.10 |
37.265 |
5.682 |
T P |
| 1eh9_A |
557 |
256 |
127 |
2.05 |
10.24 |
37.205 |
5.897 |
T P |
| 2vr5_A |
715 |
256 |
128 |
2.10 |
11.72 |
36.761 |
5.823 |
T P |
| 1m7x_B |
591 |
256 |
129 |
2.19 |
8.53 |
33.861 |
5.638 |
T P |
| 1mf7_A |
194 |
256 |
75 |
2.41 |
2.67 |
19.611 |
2.989 |
T P |
| 1mhp_B |
192 |
256 |
66 |
2.22 |
3.03 |
19.510 |
2.839 |
T P |
| 1jlm_A |
187 |
256 |
74 |
2.36 |
2.70 |
19.486 |
3.006 |
T P |
| 1shu_X |
181 |
256 |
67 |
2.36 |
2.99 |
19.452 |
2.719 |
T P |
| 2b2x_A |
188 |
256 |
65 |
2.31 |
1.54 |
19.078 |
2.698 |
T P |
| 1n3y_A |
189 |
256 |
69 |
2.37 |
5.80 |
18.936 |
2.792 |
T P |
| 1tzn_a |
181 |
256 |
71 |
2.43 |
2.82 |
18.718 |
2.811 |
T P |
| 1sht_X |
177 |
256 |
64 |
2.36 |
3.12 |
18.265 |
2.606 |
T P |
| 1m1u_A |
184 |
256 |
68 |
2.44 |
4.41 |
18.101 |
2.674 |
T P |
| 2is6_A |
654 |
256 |
65 |
2.37 |
6.15 |
17.800 |
2.637 |
T P |
| 1t6b_Y |
170 |
256 |
68 |
2.50 |
1.47 |
17.576 |
2.619 |
T P |
| 1ido_A |
184 |
256 |
66 |
2.48 |
6.06 |
17.265 |
2.559 |
T P |
| 2ok5_A |
710 |
256 |
66 |
2.51 |
3.03 |
17.211 |
2.530 |
T P |
| 1pt6_B |
195 |
256 |
68 |
2.60 |
1.47 |
17.201 |
2.519 |
T P |
| 1lsj_A |
291 |
256 |
67 |
2.70 |
4.48 |
17.112 |
2.389 |
T P |
| 1mjn_A |
179 |
256 |
66 |
2.42 |
6.06 |
17.074 |
2.619 |
T P |
| 1qhh_A |
164 |
256 |
59 |
2.33 |
11.86 |
16.770 |
2.424 |
T P |
| 1il0_A |
291 |
256 |
62 |
2.49 |
11.29 |
16.670 |
2.390 |
T P |
| 2odp_A |
490 |
256 |
63 |
2.61 |
3.17 |
16.573 |
2.322 |
T P |
| 1xuo_A |
181 |
256 |
61 |
2.50 |
6.56 |
16.564 |
2.342 |
T P |
| 1qc5_A |
192 |
256 |
65 |
2.52 |
1.54 |
16.467 |
2.478 |
T P |
| 1qcy_A |
193 |
256 |
66 |
2.59 |
1.52 |
16.356 |
2.457 |
T P |
| 2i6q_A |
503 |
256 |
62 |
2.49 |
1.61 |
16.288 |
2.393 |
T P |
| 1lso_A |
291 |
256 |
64 |
2.60 |
9.38 |
16.005 |
2.371 |
T P |
| 1ck4_B |
194 |
256 |
63 |
2.59 |
1.59 |
15.976 |
2.344 |
T P |
| 2orw_B |
173 |
256 |
56 |
2.46 |
8.93 |
15.952 |
2.186 |
T P |
| 1w36_B |
1158 |
256 |
58 |
2.50 |
1.72 |
15.923 |
2.232 |
T P |
| 3had_A |
293 |
256 |
64 |
2.76 |
1.56 |
15.848 |
2.240 |
T P |
| 1f0y_A |
291 |
256 |
62 |
2.68 |
1.61 |
15.691 |
2.227 |
T P |
| 2iue_A |
212 |
256 |
62 |
2.78 |
0.00 |
15.641 |
2.152 |
T P |
| 1qg8_A |
238 |
256 |
61 |
2.46 |
6.56 |
15.624 |
2.381 |
T P |
| 3hdh_A |
291 |
256 |
61 |
2.76 |
3.28 |
15.587 |
2.135 |
T P |
| 1pjr_A |
623 |
256 |
60 |
2.66 |
8.33 |
15.579 |
2.173 |
T P |
| 1m76_A |
291 |
256 |
63 |
2.65 |
1.59 |
15.552 |
2.290 |
T P |
| 1m75_A |
291 |
256 |
60 |
2.62 |
1.67 |
15.405 |
2.209 |
T P |
| 2adf_A |
189 |
256 |
60 |
2.81 |
5.00 |
15.314 |
2.059 |
T P |
| 2ica_A |
179 |
256 |
56 |
2.41 |
12.50 |
15.173 |
2.231 |
T P |
| 1atz_B |
189 |
256 |
54 |
2.44 |
3.70 |
15.134 |
2.128 |
T P |
| 3bqn_B |
182 |
256 |
56 |
2.61 |
10.71 |
15.126 |
2.065 |
T P |
| 1na5_A |
195 |
256 |
60 |
2.68 |
11.67 |
15.082 |
2.159 |
T P |
| 1qc5_B |
190 |
256 |
58 |
2.74 |
5.17 |
15.055 |
2.045 |
T P |
| 2hdh_A |
293 |
256 |
62 |
2.75 |
4.84 |
15.011 |
2.176 |
T P |
| 1bho_1 |
189 |
256 |
58 |
2.62 |
8.62 |
14.806 |
2.129 |
T P |
| 1f17_A |
293 |
256 |
57 |
2.76 |
3.51 |
14.791 |
1.995 |
T P |
| 1ao3_A |
187 |
256 |
52 |
2.50 |
5.77 |
14.786 |
2.004 |
T P |
| 3ckj_A |
300 |
256 |
57 |
2.76 |
5.26 |
14.777 |
1.991 |
T P |
| 1rd4_A |
184 |
256 |
57 |
2.84 |
12.28 |
14.613 |
1.938 |
T P |
| 1lfa_A |
183 |
256 |
60 |
2.81 |
6.67 |
14.528 |
2.059 |
T P |
| 1uaa_A |
636 |
256 |
56 |
2.69 |
3.57 |
14.495 |
2.009 |
T P |
| 2ffu_A |
494 |
256 |
60 |
2.79 |
6.67 |
14.306 |
2.076 |
T P |
| 1n9z_A |
184 |
256 |
53 |
2.61 |
11.32 |
14.214 |
1.954 |
T P |
| 2pjr_F |
544 |
256 |
57 |
2.75 |
8.77 |
14.151 |
2.002 |
T P |
| 1fe8_C |
191 |
256 |
56 |
2.63 |
5.36 |
14.127 |
2.054 |
T P |
| 3e26_A |
274 |
256 |
54 |
2.67 |
3.70 |
14.121 |
1.949 |
T P |
| 1t0p_A |
174 |
256 |
51 |
2.37 |
11.76 |
14.050 |
2.062 |
T P |
| 2z87_A |
601 |
256 |
54 |
2.71 |
3.70 |
13.826 |
1.920 |
T P |
| 3bn3_A |
180 |
256 |
51 |
2.65 |
15.69 |
13.749 |
1.853 |
T P |
| 2z86_C |
603 |
256 |
50 |
2.49 |
4.00 |
13.675 |
1.927 |
T P |
| 2q7s_B |
280 |
256 |
55 |
2.64 |
1.82 |
13.455 |
2.011 |
T P |
| 3e2m_A |
182 |
256 |
51 |
2.77 |
7.84 |
12.968 |
1.776 |
T P |
| 1mq9_A |
173 |
256 |
43 |
2.65 |
6.98 |
12.668 |
1.561 |
T P |
| 1qhh_B |
261 |
256 |
48 |
2.67 |
8.33 |
12.372 |
1.735 |
T P |
| 2d7i_A |
536 |
256 |
49 |
2.81 |
6.12 |
12.184 |
1.685 |
T P |
| 1mq8_B |
177 |
256 |
40 |
2.37 |
5.00 |
11.991 |
1.618 |
T P |
| 3bcv_A |
196 |
256 |
44 |
2.93 |
2.27 |
11.088 |
1.451 |
T P |
| 1xhb_A |
447 |
256 |
44 |
2.97 |
4.55 |
10.861 |
1.435 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

