LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_267.5wLII_11233_31
Total number of 3D structures: 79
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
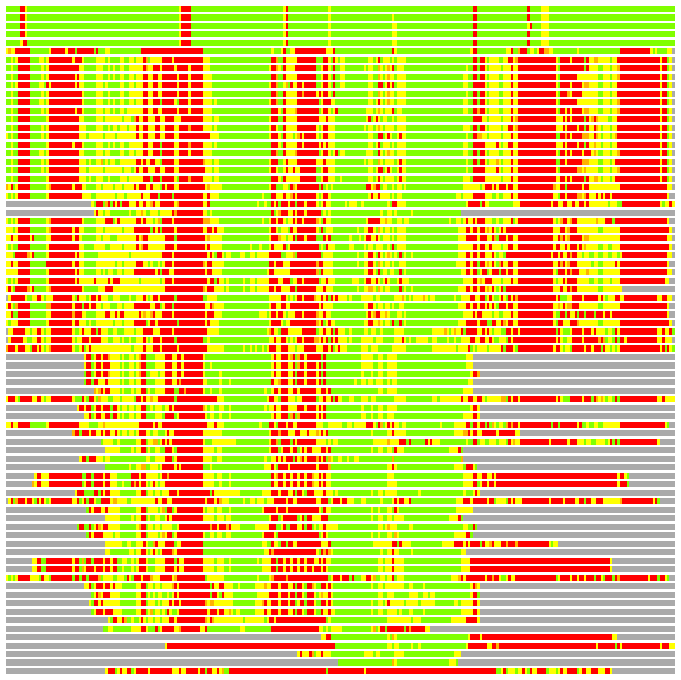
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 1oq4_A |
346 |
282 |
272 |
0.56 |
19.12 |
95.788 |
41.122 |
T P |
| 1afr_A |
345 |
282 |
272 |
0.63 |
19.12 |
95.684 |
37.057 |
T P |
| 2j2f_A |
348 |
282 |
272 |
0.63 |
19.12 |
95.659 |
37.009 |
T P |
| 2uw1_B |
338 |
282 |
272 |
0.69 |
16.91 |
95.539 |
34.225 |
T P |
| 2uw1_A |
327 |
282 |
272 |
0.74 |
16.91 |
95.438 |
32.387 |
T P |
| 1za0_A |
242 |
282 |
195 |
1.63 |
15.90 |
64.821 |
11.291 |
T P |
| 1piy_A |
340 |
282 |
179 |
2.17 |
12.29 |
46.049 |
7.886 |
T P |
| 1rib_A |
340 |
282 |
177 |
2.22 |
11.30 |
45.205 |
7.638 |
T P |
| 1piz_A |
340 |
282 |
184 |
2.28 |
11.41 |
44.831 |
7.736 |
T P |
| 1pj0_A |
340 |
282 |
178 |
2.24 |
11.80 |
44.320 |
7.615 |
T P |
| 1mxr_A |
339 |
282 |
181 |
2.28 |
10.50 |
43.659 |
7.596 |
T P |
| 1pm2_A |
339 |
282 |
180 |
2.27 |
11.67 |
43.556 |
7.602 |
T P |
| 1rnr_A |
339 |
282 |
180 |
2.31 |
11.11 |
43.483 |
7.461 |
T P |
| 1pfr_A |
340 |
282 |
179 |
2.25 |
12.29 |
43.476 |
7.602 |
T P |
| 2av8_A |
340 |
282 |
181 |
2.29 |
11.05 |
43.133 |
7.573 |
T P |
| 1rsr_B |
341 |
282 |
178 |
2.33 |
10.67 |
43.004 |
7.317 |
T P |
| 1yfd_B |
341 |
282 |
177 |
2.30 |
11.86 |
42.943 |
7.386 |
T P |
| 1biq_A |
339 |
282 |
178 |
2.26 |
10.67 |
42.847 |
7.554 |
T P |
| 1av8_A |
340 |
282 |
179 |
2.35 |
10.61 |
42.836 |
7.315 |
T P |
| 2alx_A |
339 |
282 |
177 |
2.35 |
10.73 |
42.828 |
7.221 |
T P |
| 1biq_B |
341 |
282 |
176 |
2.27 |
10.80 |
42.501 |
7.424 |
T P |
| 2rcc_A |
313 |
282 |
172 |
2.27 |
12.21 |
42.246 |
7.266 |
T P |
| 1uzr_B |
288 |
282 |
168 |
2.33 |
10.12 |
42.115 |
6.907 |
T P |
| 2oc5_A |
222 |
282 |
156 |
2.21 |
12.82 |
41.715 |
6.768 |
T P |
| 2qqy_A |
139 |
282 |
128 |
1.86 |
15.62 |
41.064 |
6.518 |
T P |
| 3dhz_B |
296 |
282 |
171 |
2.46 |
10.53 |
41.008 |
6.676 |
T P |
| 2vux_A |
272 |
282 |
169 |
2.32 |
12.43 |
40.854 |
6.997 |
T P |
| 1h0o_A |
288 |
282 |
172 |
2.39 |
11.05 |
40.745 |
6.920 |
T P |
| 1syy_A |
317 |
282 |
177 |
2.41 |
14.69 |
40.663 |
7.047 |
T P |
| 2uw2_A |
275 |
282 |
172 |
2.41 |
10.47 |
40.519 |
6.859 |
T P |
| 1jk0_A |
334 |
282 |
170 |
2.37 |
11.18 |
40.449 |
6.889 |
T P |
| 2o1z_A |
288 |
282 |
163 |
2.25 |
11.04 |
40.241 |
6.941 |
T P |
| 2ani_A |
317 |
282 |
172 |
2.44 |
14.53 |
39.912 |
6.769 |
T P |
| 2iyh_A |
275 |
282 |
170 |
2.46 |
9.41 |
39.728 |
6.630 |
T P |
| 2inn_A |
496 |
282 |
174 |
2.47 |
10.34 |
39.545 |
6.777 |
T P |
| 1smq_A |
329 |
282 |
161 |
2.40 |
11.18 |
39.416 |
6.448 |
T P |
| 1r2f_A |
283 |
282 |
163 |
2.37 |
12.88 |
39.332 |
6.596 |
T P |
| 2p1i_A |
258 |
282 |
162 |
2.31 |
11.73 |
39.305 |
6.717 |
T P |
| 2rdb_A |
491 |
282 |
170 |
2.44 |
9.41 |
39.207 |
6.684 |
T P |
| 2inp_A |
494 |
282 |
170 |
2.45 |
9.41 |
39.086 |
6.665 |
T P |
| 2inc_A |
491 |
282 |
173 |
2.56 |
9.83 |
38.934 |
6.503 |
T P |
| 2fkz_A |
155 |
282 |
129 |
1.93 |
15.50 |
38.783 |
6.355 |
T P |
| 1sof_A |
155 |
282 |
129 |
1.94 |
15.50 |
38.626 |
6.312 |
T P |
| 2vxi_A |
157 |
282 |
130 |
1.92 |
11.54 |
38.499 |
6.447 |
T P |
| 3b3h_A |
161 |
282 |
130 |
1.95 |
15.38 |
38.420 |
6.346 |
T P |
| 1nfv_B |
170 |
282 |
125 |
1.85 |
14.40 |
38.334 |
6.411 |
T P |
| 1t0q_A |
491 |
282 |
168 |
2.46 |
10.71 |
38.268 |
6.560 |
T P |
| 3fvb_A |
161 |
282 |
129 |
1.99 |
16.28 |
38.087 |
6.167 |
T P |
| 1jgc_A |
160 |
282 |
127 |
1.97 |
14.17 |
37.468 |
6.147 |
T P |
| 1jk0_B |
265 |
282 |
161 |
2.42 |
11.18 |
37.277 |
6.400 |
T P |
| 3fse_B |
349 |
282 |
134 |
2.22 |
11.19 |
36.508 |
5.779 |
T P |
| 3ez0_A |
216 |
282 |
155 |
2.29 |
10.32 |
36.417 |
6.492 |
T P |
| 1yv1_A |
202 |
282 |
119 |
1.91 |
10.08 |
35.874 |
5.929 |
T P |
| 2clb_A |
169 |
282 |
130 |
1.90 |
13.85 |
35.750 |
6.507 |
T P |
| 2fzf_A |
158 |
282 |
118 |
1.78 |
16.10 |
35.662 |
6.262 |
T P |
| 1jkv_A |
266 |
282 |
135 |
2.11 |
10.37 |
35.195 |
6.111 |
T P |
| 1o9i_A |
266 |
282 |
134 |
2.11 |
11.19 |
34.891 |
6.072 |
T P |
| 1r03_A |
171 |
282 |
130 |
2.15 |
14.62 |
34.703 |
5.772 |
T P |
| 2jcd_A |
315 |
282 |
148 |
2.55 |
8.11 |
34.619 |
5.581 |
T P |
| 1lko_A |
190 |
282 |
120 |
1.94 |
10.00 |
34.411 |
5.891 |
T P |
| 1yuz_B |
202 |
282 |
119 |
1.92 |
10.08 |
34.365 |
5.890 |
T P |
| 1z6o_M |
191 |
282 |
130 |
2.21 |
12.31 |
34.181 |
5.625 |
T P |
| 1ryt_A |
190 |
282 |
119 |
1.94 |
10.08 |
34.026 |
5.828 |
T P |
| 2itb_A |
199 |
282 |
128 |
2.17 |
17.97 |
33.815 |
5.634 |
T P |
| 1vjx_A |
149 |
282 |
112 |
1.88 |
12.50 |
32.856 |
5.652 |
T P |
| 2cwl_A |
299 |
282 |
129 |
2.08 |
13.18 |
32.801 |
5.929 |
T P |
| 2v8t_A |
302 |
282 |
128 |
2.09 |
13.28 |
32.640 |
5.843 |
T P |
| 3cht_A |
301 |
282 |
136 |
2.48 |
11.03 |
32.448 |
5.276 |
T P |
| 1vlg_A |
164 |
282 |
126 |
2.25 |
12.70 |
31.813 |
5.368 |
T P |
| 1eum_A |
161 |
282 |
124 |
2.10 |
12.10 |
31.691 |
5.638 |
T P |
| 2jd6_0 |
167 |
282 |
127 |
2.33 |
13.39 |
31.033 |
5.221 |
T P |
| 1z4a_A |
164 |
282 |
121 |
2.24 |
12.40 |
30.734 |
5.164 |
T P |
| 2oh3_A |
149 |
282 |
110 |
2.31 |
14.55 |
28.933 |
4.569 |
T P |
| 3ezu_A |
336 |
282 |
85 |
2.06 |
11.76 |
23.491 |
3.944 |
T P |
| 1zpy_D |
92 |
282 |
64 |
1.56 |
10.94 |
21.868 |
3.853 |
T P |
| 1j30_A |
141 |
282 |
66 |
1.78 |
12.12 |
21.637 |
3.511 |
T P |
| 1nnq_A |
170 |
282 |
66 |
2.06 |
13.64 |
20.479 |
3.055 |
T P |
| 1mft_A |
51 |
282 |
51 |
1.34 |
13.73 |
17.368 |
3.544 |
T P |
| 1i2a_A |
212 |
282 |
53 |
2.73 |
7.55 |
11.794 |
1.874 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

