LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_292.5wLII_11238_41
Total number of 3D structures: 72
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
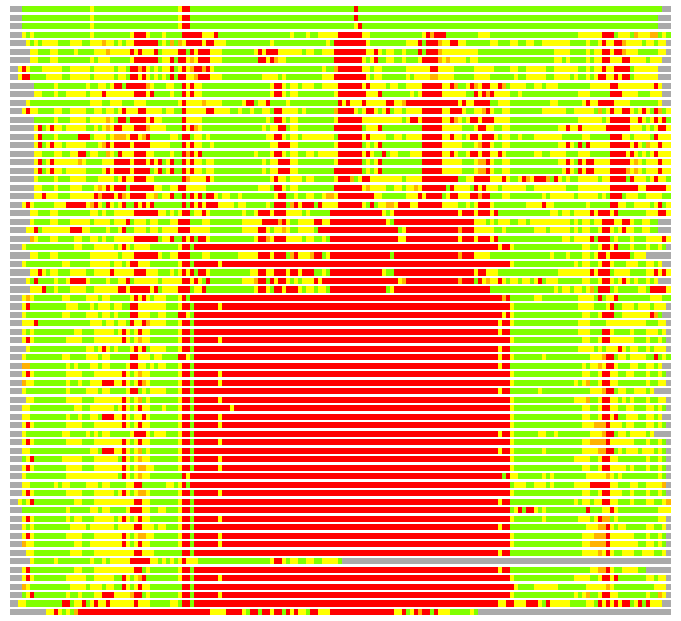
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 3bci_A |
165 |
165 |
157 |
0.56 |
17.20 |
94.649 |
23.853 |
T P |
| 3bd2_A |
164 |
165 |
156 |
0.57 |
17.31 |
94.086 |
23.363 |
T P |
| 3bck_A |
164 |
165 |
156 |
0.62 |
16.03 |
93.955 |
21.775 |
T P |
| 2in3_A |
204 |
165 |
140 |
1.97 |
13.57 |
72.313 |
6.767 |
T P |
| 3fz5_C |
192 |
165 |
131 |
2.25 |
9.92 |
59.330 |
5.570 |
T P |
| 2imd_A |
203 |
165 |
129 |
2.27 |
10.85 |
57.961 |
5.440 |
T P |
| 2imf_A |
203 |
165 |
127 |
2.12 |
9.45 |
57.846 |
5.721 |
T P |
| 1r4w_A |
221 |
165 |
138 |
2.36 |
13.77 |
56.865 |
5.612 |
T P |
| 1yzx_A |
218 |
165 |
136 |
2.35 |
13.97 |
56.328 |
5.557 |
T P |
| 1fvk_A |
188 |
165 |
126 |
2.39 |
14.29 |
53.890 |
5.070 |
T P |
| 1bq7_A |
186 |
165 |
129 |
2.23 |
9.30 |
53.884 |
5.528 |
T P |
| 1z6m_A |
175 |
165 |
127 |
2.17 |
15.75 |
52.976 |
5.604 |
T P |
| 2rem_C |
191 |
165 |
132 |
2.45 |
15.15 |
52.347 |
5.172 |
T P |
| 1bed_A |
181 |
165 |
122 |
2.28 |
9.84 |
52.342 |
5.136 |
T P |
| 2b6m_B |
188 |
165 |
123 |
2.24 |
5.69 |
51.739 |
5.254 |
T P |
| 1u3a_E |
188 |
165 |
126 |
2.38 |
10.32 |
51.607 |
5.082 |
T P |
| 1fvj_A |
188 |
165 |
119 |
2.28 |
7.56 |
50.837 |
4.995 |
T P |
| 2b3s_A |
187 |
165 |
123 |
2.34 |
10.57 |
49.907 |
5.045 |
T P |
| 1ac1_A |
188 |
165 |
119 |
2.36 |
7.56 |
49.854 |
4.845 |
T P |
| 1acv_A |
188 |
165 |
118 |
2.33 |
6.78 |
49.449 |
4.861 |
T P |
| 3c7m_A |
195 |
165 |
122 |
2.43 |
11.48 |
49.161 |
4.822 |
T P |
| 2ijy_A |
181 |
165 |
117 |
2.43 |
10.26 |
49.091 |
4.618 |
T P |
| 3feu_A |
183 |
165 |
118 |
2.34 |
17.80 |
46.675 |
4.829 |
T P |
| 2znm_C |
186 |
165 |
113 |
2.29 |
11.50 |
46.391 |
4.726 |
T P |
| 1jzd_A |
219 |
165 |
102 |
2.19 |
19.61 |
44.176 |
4.451 |
T P |
| 1v58_A |
229 |
165 |
107 |
2.37 |
15.89 |
43.146 |
4.324 |
T P |
| 2h0g_A |
229 |
165 |
109 |
2.42 |
10.09 |
43.126 |
4.324 |
T P |
| 1jzo_B |
216 |
165 |
102 |
2.25 |
21.57 |
42.730 |
4.337 |
T P |
| 2i4a_A |
107 |
165 |
77 |
1.87 |
9.09 |
41.563 |
3.907 |
T P |
| 2h0i_A |
229 |
165 |
95 |
2.19 |
16.84 |
41.343 |
4.147 |
T P |
| 2trx_A |
108 |
165 |
76 |
1.93 |
18.42 |
40.776 |
3.748 |
T P |
| 1eej_A |
216 |
165 |
100 |
2.39 |
21.00 |
40.222 |
4.020 |
T P |
| 2h0h_B |
230 |
165 |
101 |
2.39 |
8.91 |
40.192 |
4.061 |
T P |
| 1t3b_A |
209 |
165 |
95 |
2.16 |
15.79 |
39.898 |
4.213 |
T P |
| 2dj1_A |
140 |
165 |
78 |
2.05 |
17.95 |
39.872 |
3.625 |
T P |
| 3f8u_A |
469 |
165 |
77 |
2.01 |
14.29 |
39.756 |
3.644 |
T P |
| 2o8v_B |
108 |
165 |
74 |
1.98 |
16.22 |
38.971 |
3.561 |
T P |
| 1mek_A |
120 |
165 |
78 |
2.13 |
17.95 |
38.300 |
3.499 |
T P |
| 1t00_A |
112 |
165 |
77 |
1.97 |
18.18 |
38.029 |
3.724 |
T P |
| 2h6y_A |
107 |
165 |
77 |
2.05 |
19.48 |
37.738 |
3.582 |
T P |
| 2dj2_A |
120 |
165 |
76 |
2.12 |
15.79 |
37.317 |
3.419 |
T P |
| 3dyr_A |
111 |
165 |
77 |
2.00 |
15.58 |
37.305 |
3.660 |
T P |
| 1quw_A |
105 |
165 |
76 |
2.10 |
15.79 |
37.181 |
3.462 |
T P |
| 2h76_A |
108 |
165 |
77 |
2.03 |
18.18 |
37.129 |
3.623 |
T P |
| 2fch_D |
107 |
165 |
74 |
1.97 |
18.92 |
36.922 |
3.574 |
T P |
| 2eiq_A |
107 |
165 |
74 |
1.98 |
14.86 |
36.826 |
3.555 |
T P |
| 3dxb_D |
216 |
165 |
77 |
2.08 |
18.18 |
36.710 |
3.532 |
T P |
| 1txx_A |
108 |
165 |
77 |
2.04 |
18.18 |
36.702 |
3.597 |
T P |
| 2h70_A |
107 |
165 |
72 |
1.98 |
18.06 |
36.626 |
3.468 |
T P |
| 2h74_A |
108 |
165 |
77 |
2.14 |
16.88 |
36.589 |
3.443 |
T P |
| 2eir_A |
107 |
165 |
73 |
1.96 |
15.07 |
36.445 |
3.540 |
T P |
| 2h72_A |
107 |
165 |
77 |
2.16 |
16.88 |
36.432 |
3.407 |
T P |
| 2eio_A |
107 |
165 |
75 |
2.12 |
17.33 |
36.338 |
3.377 |
T P |
| 2tir_A |
108 |
165 |
77 |
2.06 |
16.88 |
35.428 |
3.564 |
T P |
| 2h6z_A |
108 |
165 |
77 |
2.09 |
18.18 |
35.407 |
3.512 |
T P |
| 1faa_A |
121 |
165 |
80 |
2.15 |
16.25 |
34.848 |
3.548 |
T P |
| 1rqm_A |
105 |
165 |
73 |
2.19 |
16.44 |
34.815 |
3.184 |
T P |
| 2h73_A |
108 |
165 |
77 |
2.07 |
19.48 |
34.262 |
3.545 |
T P |
| 1v98_A |
92 |
165 |
78 |
2.08 |
14.10 |
34.223 |
3.580 |
T P |
| 2b5e_A |
483 |
165 |
73 |
1.92 |
10.96 |
34.178 |
3.610 |
T P |
| 2h75_B |
108 |
165 |
77 |
2.12 |
18.18 |
34.127 |
3.466 |
T P |
| 1nw2_A |
105 |
165 |
78 |
2.12 |
16.67 |
34.006 |
3.510 |
T P |
| 2h71_A |
107 |
165 |
78 |
2.24 |
16.67 |
33.625 |
3.336 |
T P |
| 1zzy_A |
106 |
165 |
78 |
2.25 |
16.67 |
33.273 |
3.315 |
T P |
| 1nsw_A |
105 |
165 |
77 |
2.22 |
16.88 |
33.210 |
3.320 |
T P |
| 1dyv_A |
186 |
165 |
70 |
2.09 |
18.57 |
33.177 |
3.199 |
T P |
| 2i1u_A |
108 |
165 |
72 |
2.09 |
9.72 |
33.137 |
3.295 |
T P |
| 1keb_A |
108 |
165 |
75 |
2.09 |
17.33 |
33.062 |
3.418 |
T P |
| 2oe3_A |
106 |
165 |
77 |
2.22 |
22.08 |
33.051 |
3.320 |
T P |
| 2fd3_A |
108 |
165 |
73 |
2.00 |
17.81 |
32.416 |
3.480 |
T P |
| 1fo5_A |
85 |
165 |
64 |
2.47 |
12.50 |
24.970 |
2.491 |
T P |
| 2iyj_B |
75 |
165 |
39 |
2.72 |
10.26 |
15.300 |
1.381 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

