LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_297.5wLII_11238_53.FR_351_800
Total number of 3D structures: 57
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
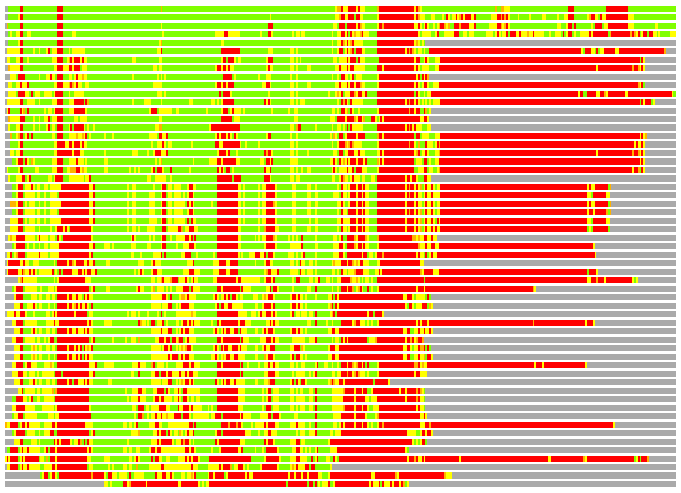
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 3cf2_A |
659 |
357 |
309 |
1.11 |
19.09 |
84.596 |
25.504 |
T P |
| 1ypw_A |
692 |
357 |
294 |
1.49 |
19.05 |
78.180 |
18.474 |
T P |
| 1r7r_A |
683 |
357 |
288 |
1.56 |
17.71 |
77.120 |
17.310 |
T P |
| 1yqi_C |
708 |
357 |
280 |
2.04 |
17.14 |
59.264 |
13.096 |
T P |
| 1s3s_F |
441 |
357 |
194 |
1.30 |
18.56 |
52.973 |
13.836 |
T P |
| 2ce7_C |
421 |
357 |
195 |
1.80 |
18.46 |
49.562 |
10.287 |
T P |
| 3b9p_A |
268 |
357 |
188 |
1.78 |
15.43 |
47.380 |
10.001 |
T P |
| 3eih_A |
319 |
357 |
194 |
1.91 |
18.56 |
47.250 |
9.665 |
T P |
| 1iy2_A |
245 |
357 |
183 |
1.73 |
18.03 |
47.131 |
10.010 |
T P |
| 3eie_A |
303 |
357 |
188 |
1.83 |
19.68 |
47.101 |
9.765 |
T P |
| 2dhr_A |
458 |
357 |
195 |
1.84 |
17.44 |
46.980 |
10.053 |
T P |
| 3d8b_A |
281 |
357 |
194 |
1.92 |
15.46 |
46.977 |
9.600 |
T P |
| 1lv7_A |
251 |
357 |
182 |
1.81 |
18.68 |
46.278 |
9.547 |
T P |
| 1ixz_A |
238 |
357 |
177 |
1.72 |
18.64 |
46.247 |
9.743 |
T P |
| 2qz4_A |
223 |
357 |
176 |
1.73 |
14.77 |
45.586 |
9.592 |
T P |
| 2qp9_X |
288 |
357 |
182 |
1.95 |
18.13 |
44.738 |
8.865 |
T P |
| 3cf0_A |
281 |
357 |
185 |
1.96 |
22.70 |
44.712 |
8.975 |
T P |
| 1xwi_A |
322 |
357 |
184 |
1.89 |
17.93 |
44.479 |
9.264 |
T P |
| 2rko_A |
280 |
357 |
180 |
1.88 |
20.00 |
44.282 |
9.073 |
T P |
| 2zan_A |
312 |
357 |
190 |
2.00 |
18.42 |
43.536 |
9.048 |
T P |
| 2r62_A |
249 |
357 |
179 |
1.88 |
17.32 |
42.557 |
9.053 |
T P |
| 1in8_A |
298 |
357 |
165 |
2.05 |
13.33 |
37.794 |
7.679 |
T P |
| 1in6_A |
300 |
357 |
165 |
2.07 |
14.55 |
37.584 |
7.606 |
T P |
| 1j7k_A |
299 |
357 |
167 |
2.12 |
13.77 |
37.480 |
7.507 |
T P |
| 1in4_A |
298 |
357 |
164 |
2.04 |
14.02 |
37.317 |
7.649 |
T P |
| 1in5_A |
301 |
357 |
165 |
2.11 |
13.33 |
37.149 |
7.480 |
T P |
| 1in7_A |
298 |
357 |
162 |
2.07 |
12.96 |
34.568 |
7.450 |
T P |
| 1ixs_B |
315 |
357 |
153 |
2.19 |
15.03 |
33.416 |
6.687 |
T P |
| 1hqc_A |
314 |
357 |
151 |
2.12 |
14.57 |
33.225 |
6.801 |
T P |
| 1ixr_C |
308 |
357 |
148 |
2.11 |
15.54 |
33.111 |
6.709 |
T P |
| 1nsf_A |
247 |
357 |
161 |
2.14 |
16.77 |
32.275 |
7.184 |
T P |
| 1d2n_A |
246 |
357 |
162 |
2.26 |
15.43 |
31.324 |
6.867 |
T P |
| 1sxj_A |
441 |
357 |
154 |
2.21 |
9.74 |
30.865 |
6.665 |
T P |
| 1sxj_C |
322 |
357 |
149 |
2.10 |
9.40 |
30.313 |
6.767 |
T P |
| 1g4a_E |
356 |
357 |
153 |
2.19 |
10.46 |
30.091 |
6.677 |
T P |
| 1ofh_A |
309 |
357 |
148 |
2.07 |
10.81 |
29.899 |
6.810 |
T P |
| 2c9o_A |
398 |
357 |
139 |
2.16 |
18.71 |
29.896 |
6.150 |
T P |
| 1sxj_D |
328 |
357 |
148 |
2.18 |
6.76 |
29.841 |
6.489 |
T P |
| 1g41_A |
334 |
357 |
149 |
2.17 |
10.74 |
29.415 |
6.552 |
T P |
| 1iqp_A |
326 |
357 |
136 |
2.20 |
12.50 |
29.402 |
5.919 |
T P |
| 1im2_A |
346 |
357 |
146 |
2.15 |
10.27 |
29.091 |
6.502 |
T P |
| 1e94_F |
408 |
357 |
151 |
2.20 |
9.27 |
29.047 |
6.572 |
T P |
| 1sxj_B |
316 |
357 |
149 |
2.30 |
10.07 |
28.701 |
6.213 |
T P |
| 1sxj_E |
317 |
357 |
143 |
2.21 |
10.49 |
28.425 |
6.190 |
T P |
| 1doo_E |
393 |
357 |
144 |
2.20 |
9.72 |
28.390 |
6.264 |
T P |
| 1xxi_C |
366 |
357 |
143 |
2.21 |
16.08 |
28.103 |
6.197 |
T P |
| 1njg_A |
240 |
357 |
143 |
2.20 |
14.69 |
27.720 |
6.213 |
T P |
| 1jr3_A |
366 |
357 |
140 |
2.21 |
14.29 |
27.710 |
6.051 |
T P |
| 2chq_A |
313 |
357 |
136 |
2.25 |
13.24 |
27.676 |
5.781 |
T P |
| 1r6b_X |
704 |
357 |
148 |
2.31 |
16.89 |
27.628 |
6.139 |
T P |
| 2chg_A |
223 |
357 |
136 |
2.12 |
12.50 |
27.573 |
6.135 |
T P |
| 1um8_A |
327 |
357 |
134 |
2.00 |
13.43 |
27.446 |
6.387 |
T P |
| 1jbk_A |
189 |
357 |
124 |
2.22 |
14.52 |
24.033 |
5.345 |
T P |
| 1qvr_A |
803 |
357 |
117 |
2.26 |
13.68 |
23.158 |
4.963 |
T P |
| 2p65_A |
185 |
357 |
125 |
2.42 |
11.20 |
22.879 |
4.964 |
T P |
| 2j37_W |
479 |
357 |
79 |
2.54 |
12.66 |
15.208 |
2.995 |
T P |
| 2di4_A |
176 |
357 |
46 |
2.51 |
4.35 |
8.604 |
1.762 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

