LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_348.5wLII_11276_155
Total number of 3D structures: 43
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
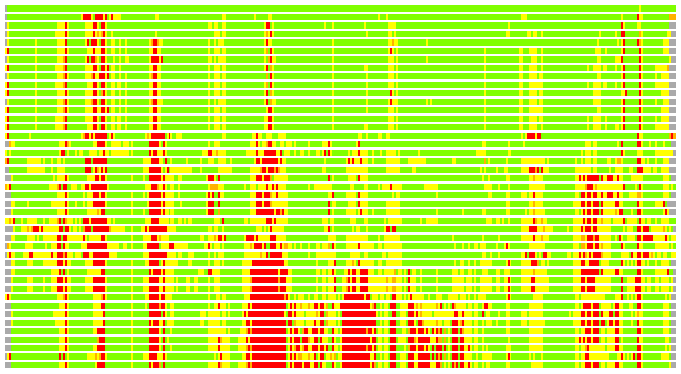
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 2d1c_A |
495 |
336 |
335 |
0.33 |
50.75 |
99.680 |
77.486 |
T P |
| 3blv_C |
344 |
336 |
324 |
1.23 |
41.36 |
92.869 |
24.321 |
T P |
| 2d4v_A |
427 |
336 |
324 |
1.24 |
38.58 |
92.266 |
24.091 |
T P |
| 1hqs_A |
423 |
336 |
327 |
1.48 |
41.28 |
91.152 |
20.652 |
T P |
| 1hj6_A |
414 |
336 |
320 |
1.37 |
38.75 |
90.397 |
21.705 |
T P |
| 6icd_A |
414 |
336 |
322 |
1.52 |
38.51 |
88.842 |
19.828 |
T P |
| 3dms_A |
413 |
336 |
319 |
1.44 |
37.30 |
88.702 |
20.734 |
T P |
| 1iso_A |
414 |
336 |
321 |
1.57 |
37.38 |
88.692 |
19.247 |
T P |
| 1gro_A |
414 |
336 |
321 |
1.56 |
38.32 |
88.476 |
19.383 |
T P |
| 1cw1_A |
415 |
336 |
322 |
1.59 |
38.51 |
88.444 |
19.038 |
T P |
| 1idd_A |
414 |
336 |
322 |
1.57 |
38.51 |
88.399 |
19.327 |
T P |
| 1bl5_A |
414 |
336 |
321 |
1.55 |
38.94 |
88.337 |
19.471 |
T P |
| 1grp_A |
414 |
336 |
320 |
1.53 |
38.75 |
88.181 |
19.629 |
T P |
| 1pb1_A |
416 |
336 |
321 |
1.56 |
38.94 |
88.081 |
19.369 |
T P |
| 1cw7_A |
415 |
336 |
321 |
1.57 |
38.63 |
88.016 |
19.203 |
T P |
| 3blx_E |
341 |
336 |
308 |
1.45 |
42.21 |
86.990 |
19.857 |
T P |
| 1a05_A |
357 |
336 |
313 |
1.62 |
37.38 |
86.547 |
18.231 |
T P |
| 1cnz_A |
363 |
336 |
314 |
1.91 |
36.31 |
81.765 |
15.649 |
T P |
| 3blx_B |
346 |
336 |
301 |
1.92 |
42.19 |
75.329 |
14.884 |
T P |
| 3blv_H |
348 |
336 |
304 |
2.03 |
40.79 |
74.774 |
14.292 |
T P |
| 1gc8_A |
345 |
336 |
294 |
1.80 |
34.69 |
73.765 |
15.488 |
T P |
| 1tyo_A |
427 |
336 |
309 |
2.06 |
37.86 |
73.348 |
14.304 |
T P |
| 1osj_A |
345 |
336 |
294 |
1.91 |
34.01 |
72.429 |
14.621 |
T P |
| 1dr8_A |
344 |
336 |
291 |
1.84 |
35.05 |
71.056 |
15.023 |
T P |
| 1dpz_A |
345 |
336 |
291 |
1.85 |
35.40 |
70.793 |
14.957 |
T P |
| 2iv0_A |
412 |
336 |
305 |
2.17 |
41.31 |
69.782 |
13.414 |
T P |
| 1wpw_A |
336 |
336 |
281 |
2.03 |
43.42 |
69.530 |
13.207 |
T P |
| 1x0l_A |
333 |
336 |
297 |
2.19 |
44.44 |
68.696 |
12.949 |
T P |
| 1vlc_A |
362 |
336 |
293 |
2.21 |
36.18 |
65.642 |
12.688 |
T P |
| 2e0c_A |
401 |
336 |
281 |
2.21 |
36.30 |
65.371 |
12.177 |
T P |
| 1v53_A |
356 |
336 |
283 |
2.06 |
35.69 |
64.916 |
13.099 |
T P |
| 1dr0_A |
346 |
336 |
278 |
2.10 |
34.53 |
63.693 |
12.629 |
T P |
| 2ayq_B |
357 |
336 |
278 |
2.25 |
37.05 |
62.003 |
11.807 |
T P |
| 1ayq_B |
357 |
336 |
278 |
2.25 |
37.05 |
62.003 |
11.807 |
T P |
| 1v5b_B |
357 |
336 |
287 |
2.05 |
34.84 |
61.183 |
13.325 |
T P |
| 1xac_A |
345 |
336 |
258 |
2.17 |
32.95 |
55.726 |
11.361 |
T P |
| 1idm_A |
343 |
336 |
259 |
2.20 |
33.59 |
55.637 |
11.279 |
T P |
| 1wal_A |
345 |
336 |
257 |
2.17 |
32.30 |
55.229 |
11.305 |
T P |
| 1g2u_A |
345 |
336 |
249 |
2.02 |
32.53 |
55.167 |
11.756 |
T P |
| 1gc9_A |
345 |
336 |
246 |
1.99 |
31.30 |
54.927 |
11.793 |
T P |
| 1ipd_A |
345 |
336 |
248 |
2.03 |
30.65 |
54.915 |
11.668 |
T P |
| 1cm7_A |
363 |
336 |
256 |
2.15 |
33.59 |
54.892 |
11.391 |
T P |
| 1xaa_A |
345 |
336 |
246 |
1.98 |
29.67 |
54.334 |
11.812 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

