LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_4.5wLII_08970_1
Total number of 3D structures: 88
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
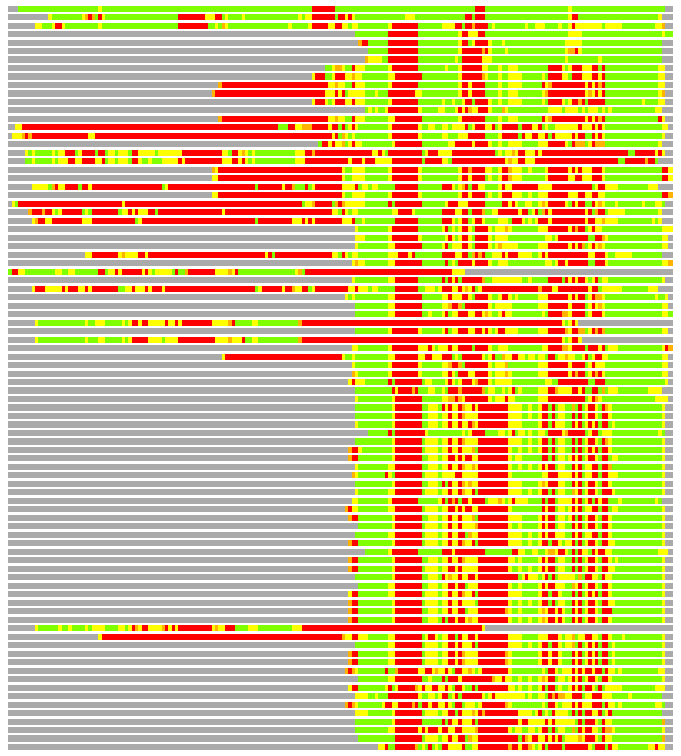
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 1m70_A |
190 |
199 |
184 |
0.42 |
16.85 |
92.193 |
35.408 |
T P |
| 1h1o_A |
172 |
199 |
155 |
1.38 |
19.35 |
74.658 |
10.505 |
T P |
| 1fcd_C |
174 |
199 |
154 |
1.93 |
13.64 |
68.747 |
7.594 |
T P |
| 1cno_G |
87 |
199 |
82 |
1.17 |
20.73 |
40.128 |
6.478 |
T P |
| 1c53_A |
79 |
199 |
74 |
1.40 |
13.51 |
35.601 |
4.937 |
T P |
| 1dvh_A |
79 |
199 |
71 |
1.45 |
19.72 |
34.960 |
4.581 |
T P |
| 2dvh_A |
79 |
199 |
74 |
1.56 |
17.57 |
34.711 |
4.445 |
T P |
| 1e2r_B |
543 |
199 |
77 |
2.00 |
16.88 |
33.345 |
3.673 |
T P |
| 1hzu_A |
521 |
199 |
76 |
2.02 |
22.37 |
32.939 |
3.588 |
T P |
| 1qks_A |
559 |
199 |
73 |
2.16 |
20.55 |
31.030 |
3.230 |
T P |
| 1hj5_A |
559 |
199 |
74 |
2.18 |
18.92 |
30.779 |
3.246 |
T P |
| 1hzv_A |
514 |
199 |
73 |
2.07 |
26.03 |
30.509 |
3.359 |
T P |
| 1e08_E |
78 |
199 |
75 |
2.07 |
18.67 |
30.270 |
3.455 |
T P |
| 1gq1_A |
559 |
199 |
71 |
2.14 |
19.72 |
29.455 |
3.163 |
T P |
| 1kv9_A |
664 |
199 |
83 |
2.35 |
15.66 |
28.991 |
3.386 |
T P |
| 1yiq_A |
684 |
199 |
78 |
2.25 |
10.26 |
28.710 |
3.320 |
T P |
| 1nir_B |
539 |
199 |
75 |
2.25 |
25.33 |
28.551 |
3.190 |
T P |
| 2c1v_A |
335 |
199 |
85 |
2.52 |
7.06 |
28.512 |
3.241 |
T P |
| 2vhd_A |
323 |
199 |
82 |
2.49 |
10.98 |
28.178 |
3.165 |
T P |
| 1mg2_D |
147 |
199 |
72 |
2.01 |
15.28 |
28.097 |
3.417 |
T P |
| 2gc4_D |
147 |
199 |
72 |
1.93 |
15.28 |
28.041 |
3.548 |
T P |
| 1iqc_A |
308 |
199 |
79 |
2.41 |
10.13 |
27.773 |
3.152 |
T P |
| 2d0w_A |
168 |
199 |
72 |
2.02 |
16.67 |
27.652 |
3.396 |
T P |
| 1kb0_A |
669 |
199 |
77 |
2.26 |
15.58 |
27.560 |
3.265 |
T P |
| 2c1d_A |
264 |
199 |
79 |
2.31 |
11.39 |
27.495 |
3.284 |
T P |
| 1zzh_B |
308 |
199 |
78 |
2.36 |
12.82 |
27.467 |
3.170 |
T P |
| 1cyj_A |
90 |
199 |
70 |
2.00 |
17.14 |
27.182 |
3.337 |
T P |
| 2v07_A |
98 |
199 |
68 |
1.93 |
13.24 |
26.921 |
3.349 |
T P |
| 1gdv_A |
85 |
199 |
70 |
2.06 |
20.00 |
26.830 |
3.244 |
T P |
| 1h32_A |
261 |
199 |
77 |
2.36 |
10.39 |
26.681 |
3.134 |
T P |
| 2ce0_A |
99 |
199 |
68 |
1.95 |
17.65 |
26.536 |
3.311 |
T P |
| 1umm_A |
149 |
199 |
71 |
2.09 |
8.45 |
26.398 |
3.247 |
T P |
| 1w2l_A |
97 |
199 |
69 |
1.97 |
20.29 |
26.267 |
3.339 |
T P |
| 1nml_A |
316 |
199 |
76 |
2.55 |
19.74 |
26.170 |
2.871 |
T P |
| 1ls9_A |
91 |
199 |
70 |
2.00 |
17.14 |
26.130 |
3.334 |
T P |
| 1f1f_A |
88 |
199 |
66 |
1.99 |
18.18 |
25.884 |
3.164 |
T P |
| 2zbo_A |
86 |
199 |
69 |
2.05 |
15.94 |
25.823 |
3.208 |
T P |
| 1dt1_A |
129 |
199 |
68 |
2.11 |
14.71 |
25.806 |
3.083 |
T P |
| 2v08_A |
84 |
199 |
68 |
2.06 |
19.12 |
25.719 |
3.149 |
T P |
| 1c52_A |
131 |
199 |
67 |
2.20 |
14.93 |
25.328 |
2.908 |
T P |
| 1c6s_A |
87 |
199 |
65 |
2.11 |
18.46 |
25.266 |
2.938 |
T P |
| 3cp5_A |
116 |
199 |
73 |
2.25 |
9.59 |
25.182 |
3.111 |
T P |
| 1kib_A |
89 |
199 |
67 |
2.02 |
17.91 |
25.162 |
3.154 |
T P |
| 1ctj_A |
89 |
199 |
68 |
2.14 |
19.12 |
25.095 |
3.041 |
T P |
| 2dge_A |
102 |
199 |
67 |
1.94 |
14.93 |
25.070 |
3.290 |
T P |
| 2c1d_B |
137 |
199 |
69 |
2.23 |
18.84 |
24.794 |
2.959 |
T P |
| 1c6r_A |
88 |
199 |
65 |
2.09 |
20.00 |
24.598 |
2.961 |
T P |
| 2b4z_A |
104 |
199 |
65 |
2.10 |
15.38 |
24.354 |
2.956 |
T P |
| 1ccr_A |
111 |
199 |
65 |
2.12 |
15.38 |
24.223 |
2.924 |
T P |
| 2b0z_B |
108 |
199 |
64 |
2.08 |
18.75 |
24.174 |
2.934 |
T P |
| 1foc_A |
128 |
199 |
63 |
1.89 |
20.63 |
24.127 |
3.167 |
T P |
| 1wej_F |
104 |
199 |
66 |
2.22 |
13.64 |
24.123 |
2.840 |
T P |
| 2ycc_A |
108 |
199 |
65 |
2.17 |
16.92 |
24.040 |
2.868 |
T P |
| 1yeb_A |
108 |
199 |
62 |
2.06 |
16.13 |
23.923 |
2.866 |
T P |
| 1irv_A |
108 |
199 |
65 |
2.15 |
16.92 |
23.901 |
2.893 |
T P |
| 1csv_A |
108 |
199 |
65 |
2.14 |
15.38 |
23.898 |
2.907 |
T P |
| 1i54_A |
103 |
199 |
64 |
2.08 |
15.62 |
23.883 |
2.933 |
T P |
| 3cx5_W |
108 |
199 |
64 |
2.14 |
17.19 |
23.849 |
2.851 |
T P |
| 1cyc_A |
103 |
199 |
66 |
2.13 |
19.70 |
23.806 |
2.958 |
T P |
| 1raq_A |
108 |
199 |
64 |
2.16 |
17.19 |
23.654 |
2.833 |
T P |
| 2b11_B |
108 |
199 |
65 |
2.24 |
16.92 |
23.579 |
2.776 |
T P |
| 1chh_A |
108 |
199 |
64 |
2.18 |
17.19 |
23.494 |
2.806 |
T P |
| 1crh_A |
108 |
199 |
64 |
2.13 |
17.19 |
23.484 |
2.871 |
T P |
| 1cri_A |
108 |
199 |
63 |
2.18 |
17.46 |
23.478 |
2.766 |
T P |
| 1wve_C |
75 |
199 |
62 |
2.00 |
25.81 |
23.419 |
2.958 |
T P |
| 1s6v_B |
108 |
199 |
65 |
2.22 |
16.92 |
23.415 |
2.801 |
T P |
| 1csu_A |
108 |
199 |
64 |
2.16 |
17.19 |
23.413 |
2.829 |
T P |
| 2aiu_A |
104 |
199 |
63 |
2.19 |
12.70 |
23.407 |
2.756 |
T P |
| 1chj_A |
108 |
199 |
63 |
2.12 |
17.46 |
23.386 |
2.836 |
T P |
| 2b12_B |
108 |
199 |
63 |
2.12 |
17.46 |
23.378 |
2.836 |
T P |
| 1cig_A |
108 |
199 |
65 |
2.20 |
16.92 |
23.373 |
2.830 |
T P |
| 1irw_A |
108 |
199 |
64 |
2.22 |
17.19 |
23.284 |
2.753 |
T P |
| 1ctz_A |
108 |
199 |
62 |
2.07 |
17.74 |
23.284 |
2.853 |
T P |
| 2fwl_A |
129 |
199 |
63 |
2.30 |
14.29 |
23.275 |
2.621 |
T P |
| 1fhb_A |
108 |
199 |
65 |
2.38 |
15.38 |
23.247 |
2.616 |
T P |
| 1csw_A |
108 |
199 |
63 |
2.09 |
15.87 |
23.240 |
2.874 |
T P |
| 1ycc_A |
108 |
199 |
64 |
2.21 |
17.19 |
23.005 |
2.768 |
T P |
| 2jqr_A |
108 |
199 |
64 |
2.21 |
17.19 |
23.005 |
2.768 |
T P |
| 1rap_A |
108 |
199 |
66 |
2.34 |
9.09 |
22.997 |
2.702 |
T P |
| 1csx_A |
108 |
199 |
62 |
2.00 |
17.74 |
22.995 |
2.954 |
T P |
| 2bcn_B |
108 |
199 |
63 |
2.24 |
11.11 |
22.772 |
2.688 |
T P |
| 1yfc_A |
108 |
199 |
65 |
2.34 |
18.46 |
22.710 |
2.665 |
T P |
| 1chi_A |
108 |
199 |
66 |
2.43 |
9.09 |
22.280 |
2.612 |
T P |
| 1j3s_A |
104 |
199 |
62 |
2.22 |
12.90 |
22.171 |
2.674 |
T P |
| 2jti_B |
103 |
199 |
60 |
2.11 |
16.67 |
22.111 |
2.710 |
T P |
| 1crg_A |
108 |
199 |
62 |
2.23 |
14.52 |
21.927 |
2.660 |
T P |
| 2b10_B |
108 |
199 |
59 |
2.28 |
18.64 |
21.080 |
2.477 |
T P |
| 1lms_A |
95 |
199 |
45 |
2.51 |
8.89 |
15.752 |
1.722 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

