LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_411.5wLII_11277_158
Total number of 3D structures: 110
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
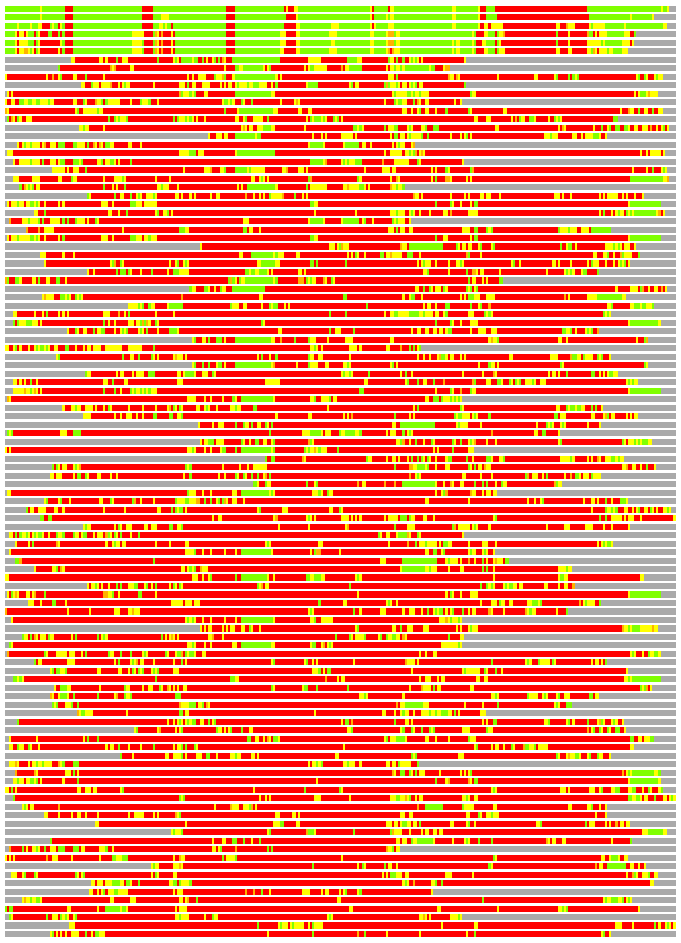
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 1p4d_B |
276 |
327 |
257 |
0.82 |
23.35 |
77.567 |
28.085 |
T P |
| 2q7t_A |
267 |
327 |
250 |
0.97 |
23.60 |
74.816 |
23.373 |
T P |
| 2a0i_A |
293 |
327 |
258 |
1.44 |
20.93 |
73.998 |
16.748 |
T P |
| 1osb_A |
287 |
327 |
227 |
1.66 |
29.52 |
66.211 |
12.919 |
T P |
| 2cdm_A |
280 |
327 |
227 |
1.66 |
29.52 |
65.096 |
12.903 |
T P |
| 1omh_A |
282 |
327 |
225 |
1.75 |
29.33 |
63.402 |
12.182 |
T P |
| 1taq_A |
807 |
327 |
76 |
2.25 |
10.53 |
17.104 |
3.236 |
T P |
| 1ega_B |
293 |
327 |
69 |
2.32 |
1.45 |
15.514 |
2.849 |
T P |
| 2zws_A |
630 |
327 |
70 |
2.57 |
5.71 |
14.713 |
2.623 |
T P |
| 1bgx_T |
828 |
327 |
76 |
2.80 |
10.53 |
14.062 |
2.616 |
T P |
| 2c03_B |
297 |
327 |
64 |
2.41 |
7.81 |
14.000 |
2.550 |
T P |
| 1goj_A |
354 |
327 |
69 |
2.70 |
1.45 |
13.736 |
2.460 |
T P |
| 1w36_B |
1158 |
327 |
62 |
2.54 |
9.68 |
13.534 |
2.347 |
T P |
| 2hko_A |
647 |
327 |
69 |
2.61 |
10.14 |
13.446 |
2.542 |
T P |
| 1gm5_A |
729 |
327 |
65 |
2.72 |
3.08 |
13.148 |
2.302 |
T P |
| 2fmp_A |
326 |
327 |
67 |
2.74 |
5.97 |
13.002 |
2.356 |
T P |
| 1f9w_A |
300 |
327 |
62 |
2.61 |
4.84 |
12.975 |
2.291 |
T P |
| 2j7p_A |
292 |
327 |
58 |
2.38 |
8.62 |
12.942 |
2.336 |
T P |
| 3kar_A |
323 |
327 |
62 |
2.40 |
0.00 |
12.775 |
2.478 |
T P |
| 2v3c_C |
403 |
327 |
60 |
2.71 |
5.00 |
12.672 |
2.138 |
T P |
| 2ffh_A |
407 |
327 |
60 |
2.66 |
8.33 |
12.595 |
2.174 |
T P |
| 1g8p_A |
321 |
327 |
64 |
2.53 |
12.50 |
12.579 |
2.434 |
T P |
| 1hei_A |
443 |
327 |
64 |
2.96 |
7.81 |
12.561 |
2.091 |
T P |
| 2j45_A |
297 |
327 |
60 |
2.32 |
5.00 |
12.478 |
2.482 |
T P |
| 2o8b_A |
831 |
327 |
60 |
2.58 |
13.33 |
12.375 |
2.239 |
T P |
| 1f9u_A |
310 |
327 |
56 |
2.40 |
5.36 |
12.366 |
2.236 |
T P |
| 1ye8_A |
172 |
327 |
58 |
2.50 |
6.90 |
12.167 |
2.232 |
T P |
| 1ls1_A |
289 |
327 |
60 |
2.37 |
5.00 |
12.131 |
2.431 |
T P |
| 2yvu_A |
179 |
327 |
57 |
2.30 |
7.02 |
12.084 |
2.376 |
T P |
| 1fnn_A |
379 |
327 |
60 |
2.58 |
8.33 |
12.034 |
2.242 |
T P |
| 2pjr_F |
544 |
327 |
58 |
2.57 |
5.17 |
11.982 |
2.171 |
T P |
| 2va8_B |
695 |
327 |
56 |
2.53 |
3.57 |
11.961 |
2.127 |
T P |
| 1pjr_A |
623 |
327 |
55 |
2.43 |
7.27 |
11.895 |
2.173 |
T P |
| 2e57_A |
606 |
327 |
53 |
2.45 |
5.66 |
11.885 |
2.081 |
T P |
| 2i3b_A |
189 |
327 |
58 |
2.47 |
6.90 |
11.779 |
2.261 |
T P |
| 1a1v_A |
432 |
327 |
58 |
2.59 |
8.62 |
11.776 |
2.154 |
T P |
| 2o8e_A |
824 |
327 |
55 |
2.65 |
7.27 |
11.702 |
2.004 |
T P |
| 1ffh_A |
287 |
327 |
53 |
2.38 |
3.77 |
11.688 |
2.139 |
T P |
| 2ce7_C |
421 |
327 |
63 |
2.86 |
4.76 |
11.625 |
2.131 |
T P |
| 2h58_A |
316 |
327 |
53 |
2.45 |
7.55 |
11.589 |
2.075 |
T P |
| 1ry6_A |
319 |
327 |
59 |
2.70 |
5.08 |
11.585 |
2.104 |
T P |
| 2is6_A |
654 |
327 |
57 |
2.72 |
3.51 |
11.492 |
2.018 |
T P |
| 1f9v_A |
313 |
327 |
52 |
2.44 |
5.77 |
11.435 |
2.043 |
T P |
| 2f55_A |
432 |
327 |
55 |
2.68 |
5.45 |
11.403 |
1.978 |
T P |
| 2iw5_A |
666 |
327 |
56 |
2.78 |
0.00 |
11.388 |
1.944 |
T P |
| 1jpn_A |
296 |
327 |
54 |
2.36 |
3.70 |
11.382 |
2.199 |
T P |
| 1fts_A |
295 |
327 |
53 |
2.32 |
9.43 |
11.319 |
2.188 |
T P |
| 2z83_A |
426 |
327 |
55 |
2.81 |
3.64 |
11.312 |
1.888 |
T P |
| 2zjo_A |
444 |
327 |
55 |
2.77 |
3.64 |
11.251 |
1.915 |
T P |
| 1zu4_A |
305 |
327 |
51 |
2.32 |
5.88 |
11.194 |
2.110 |
T P |
| 1xzp_A |
456 |
327 |
49 |
2.34 |
2.04 |
10.972 |
2.007 |
T P |
| 2zzz_A |
186 |
327 |
56 |
2.59 |
8.93 |
10.869 |
2.085 |
T P |
| 2eww_A |
343 |
327 |
47 |
2.29 |
8.51 |
10.762 |
1.967 |
T P |
| 1l8q_A |
321 |
327 |
51 |
2.76 |
5.88 |
10.692 |
1.786 |
T P |
| 1sxj_D |
328 |
327 |
52 |
2.82 |
5.77 |
10.674 |
1.783 |
T P |
| 1e69_A |
263 |
327 |
55 |
2.74 |
9.09 |
10.562 |
1.938 |
T P |
| 2hcb_D |
320 |
327 |
47 |
2.41 |
2.13 |
10.562 |
1.876 |
T P |
| 2px0_A |
258 |
327 |
47 |
2.44 |
10.64 |
10.557 |
1.848 |
T P |
| 1ihu_A |
540 |
327 |
52 |
2.72 |
1.92 |
10.531 |
1.847 |
T P |
| 2csd_A |
516 |
327 |
50 |
2.83 |
8.00 |
10.500 |
1.708 |
T P |
| 2eyq_A |
1146 |
327 |
50 |
2.74 |
4.00 |
10.477 |
1.758 |
T P |
| 8ohm_A |
435 |
327 |
52 |
2.81 |
7.69 |
10.409 |
1.784 |
T P |
| 1i5s_A |
330 |
327 |
51 |
2.61 |
5.88 |
10.372 |
1.879 |
T P |
| 1qvr_A |
803 |
327 |
50 |
2.86 |
12.00 |
10.260 |
1.689 |
T P |
| 1rj9_B |
282 |
327 |
45 |
2.34 |
8.89 |
10.186 |
1.846 |
T P |
| 1sxj_C |
322 |
327 |
42 |
2.27 |
11.90 |
10.120 |
1.776 |
T P |
| 3dkp_A |
240 |
327 |
47 |
2.60 |
2.13 |
10.081 |
1.739 |
T P |
| 3dzd_A |
368 |
327 |
44 |
2.35 |
13.64 |
10.079 |
1.794 |
T P |
| 1w36_D |
538 |
327 |
49 |
2.72 |
10.20 |
10.043 |
1.738 |
T P |
| 3dm5_A |
416 |
327 |
44 |
2.49 |
18.18 |
10.018 |
1.696 |
T P |
| 2sqc_A |
623 |
327 |
50 |
2.73 |
4.00 |
10.001 |
1.769 |
T P |
| 2qby_A |
366 |
327 |
48 |
2.91 |
8.33 |
9.920 |
1.597 |
T P |
| 3dm9_B |
305 |
327 |
47 |
2.45 |
6.38 |
9.918 |
1.842 |
T P |
| 3a00_A |
153 |
327 |
49 |
2.89 |
8.16 |
9.868 |
1.637 |
T P |
| 2vou_A |
393 |
327 |
50 |
2.67 |
2.00 |
9.859 |
1.808 |
T P |
| 3e70_C |
307 |
327 |
45 |
2.41 |
6.67 |
9.721 |
1.793 |
T P |
| 1c4o_A |
504 |
327 |
48 |
2.70 |
2.08 |
9.593 |
1.717 |
T P |
| 1s2m_A |
377 |
327 |
49 |
2.89 |
10.20 |
9.552 |
1.639 |
T P |
| 2awn_B |
374 |
327 |
46 |
2.66 |
2.17 |
9.524 |
1.667 |
T P |
| 1vma_A |
294 |
327 |
43 |
2.41 |
16.28 |
9.475 |
1.712 |
T P |
| 1g29_1 |
372 |
327 |
44 |
2.77 |
6.82 |
9.460 |
1.531 |
T P |
| 3e1s_A |
517 |
327 |
44 |
2.65 |
9.09 |
9.438 |
1.601 |
T P |
| 2pl3_A |
232 |
327 |
45 |
2.53 |
6.67 |
9.256 |
1.711 |
T P |
| 1njg_A |
240 |
327 |
42 |
2.59 |
2.38 |
9.221 |
1.563 |
T P |
| 1cu1_A |
645 |
327 |
43 |
2.63 |
4.65 |
9.204 |
1.574 |
T P |
| 2gsz_A |
343 |
327 |
44 |
2.56 |
2.27 |
9.095 |
1.656 |
T P |
| 2bgw_A |
219 |
327 |
42 |
2.62 |
4.76 |
9.009 |
1.545 |
T P |
| 2dw4_A |
634 |
327 |
44 |
2.85 |
6.82 |
8.981 |
1.490 |
T P |
| 2bmf_B |
443 |
327 |
47 |
2.89 |
8.51 |
8.873 |
1.571 |
T P |
| 1bg2_A |
323 |
327 |
46 |
2.90 |
6.52 |
8.868 |
1.534 |
T P |
| 1np6_B |
169 |
327 |
41 |
2.59 |
2.44 |
8.751 |
1.523 |
T P |
| 2ng1_A |
293 |
327 |
38 |
2.24 |
5.26 |
8.699 |
1.627 |
T P |
| 1uaa_A |
636 |
327 |
42 |
2.77 |
4.76 |
8.691 |
1.461 |
T P |
| 3kin_B |
117 |
327 |
45 |
2.67 |
2.22 |
8.643 |
1.624 |
T P |
| 2pt7_A |
323 |
327 |
42 |
2.64 |
9.52 |
8.516 |
1.535 |
T P |
| 3bh1_D |
487 |
327 |
40 |
3.03 |
5.00 |
8.437 |
1.280 |
T P |
| 1d2m_A |
552 |
327 |
42 |
2.94 |
11.90 |
8.421 |
1.381 |
T P |
| 2kin_B |
100 |
327 |
40 |
2.58 |
10.00 |
8.269 |
1.495 |
T P |
| 2zzy_A |
186 |
327 |
39 |
2.57 |
10.26 |
8.259 |
1.458 |
T P |
| 2gry_A |
327 |
327 |
37 |
2.67 |
5.41 |
8.229 |
1.336 |
T P |
| 2ewv_A |
343 |
327 |
39 |
2.58 |
12.82 |
8.211 |
1.456 |
T P |
| 1vdd_A |
199 |
327 |
35 |
2.61 |
2.86 |
8.073 |
1.292 |
T P |
| 2bhn_A |
211 |
327 |
40 |
2.64 |
0.00 |
7.967 |
1.460 |
T P |
| 1qhh_A |
164 |
327 |
38 |
2.57 |
5.26 |
7.780 |
1.421 |
T P |
| 2imj_A |
158 |
327 |
36 |
3.05 |
11.11 |
7.663 |
1.144 |
T P |
| 2v1c_A |
198 |
327 |
36 |
2.64 |
2.78 |
7.434 |
1.316 |
T P |
| 2eyu_A |
247 |
327 |
33 |
2.55 |
9.09 |
7.309 |
1.245 |
T P |
| 2qy9_A |
300 |
327 |
37 |
2.91 |
13.51 |
7.168 |
1.231 |
T P |
| 2gk6_A |
602 |
327 |
35 |
2.87 |
8.57 |
6.846 |
1.178 |
T P |
| 2iyk_A |
155 |
327 |
27 |
2.80 |
3.70 |
5.709 |
0.932 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

