LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_414.5wLII_11277_179
Total number of 3D structures: 45
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
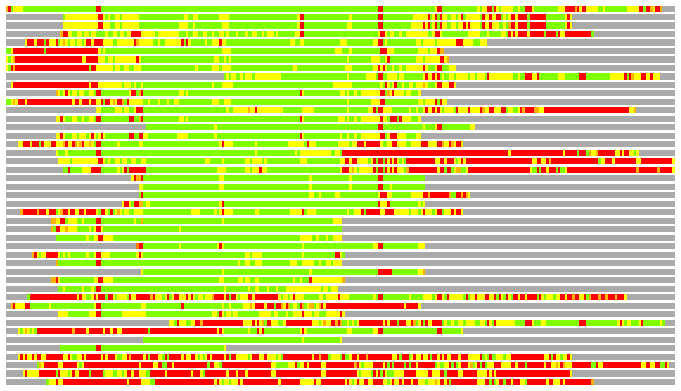
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 1w3b_A |
388 |
282 |
251 |
1.37 |
14.74 |
85.447 |
17.069 |
T P |
| 2fi7_A |
223 |
282 |
187 |
1.96 |
11.76 |
57.425 |
9.090 |
T P |
| 2ho1_A |
222 |
282 |
187 |
2.01 |
11.76 |
56.302 |
8.847 |
T P |
| 2vq2_A |
220 |
282 |
185 |
2.07 |
11.89 |
53.076 |
8.537 |
T P |
| 2q7f_A |
194 |
282 |
172 |
2.18 |
12.79 |
49.039 |
7.541 |
T P |
| 3cv0_A |
300 |
282 |
146 |
1.60 |
10.96 |
48.907 |
8.601 |
T P |
| 2c0m_C |
302 |
282 |
150 |
1.82 |
14.67 |
48.001 |
7.821 |
T P |
| 2c0l_A |
292 |
282 |
151 |
1.90 |
14.57 |
47.851 |
7.565 |
T P |
| 2pl2_A |
194 |
282 |
166 |
2.12 |
10.84 |
47.488 |
7.486 |
T P |
| 3cvq_A |
289 |
282 |
147 |
1.86 |
10.88 |
47.195 |
7.501 |
T P |
| 1e96_B |
185 |
282 |
144 |
1.78 |
7.64 |
47.082 |
7.647 |
T P |
| 2j9q_A |
300 |
282 |
147 |
1.84 |
15.65 |
46.304 |
7.577 |
T P |
| 1xnf_B |
262 |
282 |
170 |
2.26 |
9.41 |
46.201 |
7.202 |
T P |
| 1hh8_A |
192 |
282 |
142 |
1.74 |
8.45 |
46.101 |
7.714 |
T P |
| 2fo7_A |
136 |
282 |
135 |
1.10 |
11.11 |
46.031 |
11.254 |
T P |
| 1wm5_A |
205 |
282 |
141 |
1.82 |
7.09 |
45.346 |
7.340 |
T P |
| 3edt_B |
258 |
282 |
153 |
2.21 |
11.11 |
44.718 |
6.621 |
T P |
| 1wao_1 |
471 |
282 |
134 |
1.79 |
9.70 |
42.506 |
7.095 |
T P |
| 2vsy_A |
547 |
282 |
154 |
2.32 |
15.58 |
42.441 |
6.361 |
T P |
| 2vsn_A |
534 |
282 |
144 |
2.02 |
16.67 |
41.812 |
6.808 |
T P |
| 1p5q_A |
283 |
282 |
120 |
1.25 |
8.33 |
41.590 |
8.872 |
T P |
| 1qz2_A |
285 |
282 |
119 |
1.14 |
8.40 |
41.250 |
9.605 |
T P |
| 2c2l_A |
281 |
282 |
123 |
1.68 |
9.76 |
41.107 |
6.912 |
T P |
| 1ihg_A |
364 |
282 |
121 |
1.41 |
9.09 |
41.026 |
8.023 |
T P |
| 3ceq_B |
269 |
282 |
143 |
2.22 |
9.79 |
40.676 |
6.166 |
T P |
| 1kt1_A |
374 |
282 |
119 |
1.54 |
12.61 |
39.894 |
7.266 |
T P |
| 1na0_A |
119 |
282 |
119 |
1.44 |
15.13 |
39.688 |
7.743 |
T P |
| 2vyi_A |
128 |
282 |
119 |
1.39 |
15.13 |
39.645 |
7.986 |
T P |
| 2fbn_A |
153 |
282 |
117 |
1.40 |
11.97 |
39.603 |
7.813 |
T P |
| 2dba_A |
148 |
282 |
121 |
1.61 |
15.70 |
39.307 |
7.071 |
T P |
| 1a17_A |
159 |
282 |
119 |
1.49 |
10.92 |
39.156 |
7.462 |
T P |
| 1kt0_A |
357 |
282 |
114 |
1.13 |
9.65 |
39.102 |
9.233 |
T P |
| 2bug_A |
131 |
282 |
120 |
1.85 |
10.83 |
38.887 |
6.138 |
T P |
| 1elw_A |
117 |
282 |
116 |
1.63 |
14.66 |
38.054 |
6.699 |
T P |
| 2gw1_A |
487 |
282 |
153 |
2.50 |
3.92 |
37.902 |
5.879 |
T P |
| 2e2e_A |
171 |
282 |
117 |
1.97 |
10.26 |
37.314 |
5.653 |
T P |
| 1elr_A |
128 |
282 |
116 |
2.00 |
12.07 |
36.322 |
5.524 |
T P |
| 1fch_A |
302 |
282 |
137 |
2.28 |
9.49 |
35.406 |
5.749 |
T P |
| 2if4_A |
258 |
282 |
111 |
1.77 |
6.31 |
35.032 |
5.950 |
T P |
| 1na3_A |
86 |
282 |
84 |
0.88 |
14.29 |
29.369 |
8.546 |
T P |
| 2avp_A |
68 |
282 |
68 |
0.84 |
19.12 |
23.813 |
7.244 |
T P |
| 1ouv_A |
265 |
282 |
109 |
2.74 |
8.26 |
23.107 |
3.844 |
T P |
| 1xi4_A |
1630 |
282 |
91 |
2.71 |
15.38 |
20.945 |
3.239 |
T P |
| 1o5m_A |
315 |
282 |
77 |
2.75 |
12.99 |
18.526 |
2.701 |
T P |
| 1o1t_A |
315 |
282 |
76 |
2.95 |
6.58 |
18.103 |
2.489 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

