LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_432.5wLII_11277_252
Total number of 3D structures: 55
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
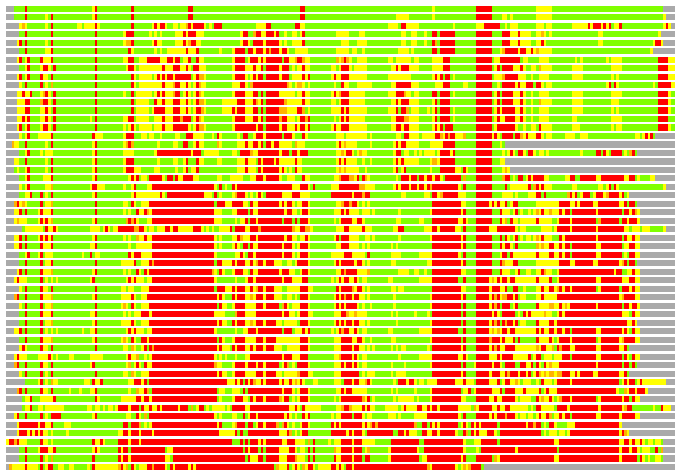
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 1wcv_1 |
243 |
257 |
237 |
0.68 |
24.47 |
91.225 |
30.396 |
T P |
| 2bek_A |
244 |
257 |
237 |
1.12 |
24.05 |
88.938 |
19.493 |
T P |
| 2oze_A |
284 |
257 |
217 |
1.82 |
19.35 |
66.731 |
11.311 |
T P |
| 1g3q_A |
237 |
257 |
210 |
1.69 |
27.62 |
66.411 |
11.760 |
T P |
| 1ion_A |
243 |
257 |
208 |
1.72 |
26.92 |
65.558 |
11.443 |
T P |
| 1hyq_A |
232 |
257 |
201 |
1.81 |
30.35 |
61.966 |
10.511 |
T P |
| 1g20_H |
262 |
257 |
195 |
1.98 |
21.54 |
57.292 |
9.374 |
T P |
| 2afh_E |
289 |
257 |
197 |
2.00 |
18.78 |
56.762 |
9.400 |
T P |
| 2c8v_A |
271 |
257 |
196 |
1.96 |
21.94 |
56.731 |
9.496 |
T P |
| 3end_A |
268 |
257 |
194 |
2.00 |
18.04 |
56.597 |
9.256 |
T P |
| 1de0_A |
289 |
257 |
199 |
2.10 |
19.60 |
56.500 |
9.063 |
T P |
| 1xdb_A |
289 |
257 |
198 |
2.08 |
19.70 |
55.282 |
9.090 |
T P |
| 1xd8_A |
289 |
257 |
196 |
2.06 |
19.39 |
54.812 |
9.062 |
T P |
| 1cp2_A |
269 |
257 |
195 |
2.06 |
20.51 |
54.194 |
9.008 |
T P |
| 1rw4_A |
271 |
257 |
186 |
1.99 |
20.97 |
52.836 |
8.886 |
T P |
| 3ea0_B |
242 |
257 |
195 |
2.08 |
23.59 |
52.632 |
8.954 |
T P |
| 2ved_B |
254 |
257 |
160 |
1.97 |
21.25 |
51.708 |
7.744 |
T P |
| 2ph1_A |
247 |
257 |
172 |
1.96 |
25.00 |
49.487 |
8.348 |
T P |
| 3bfv_A |
241 |
257 |
157 |
1.90 |
21.02 |
48.876 |
7.843 |
T P |
| 3cio_A |
255 |
257 |
158 |
1.83 |
17.09 |
48.272 |
8.174 |
T P |
| 3fkq_A |
354 |
257 |
163 |
2.00 |
15.95 |
45.453 |
7.744 |
T P |
| 3cwq_A |
206 |
257 |
154 |
1.96 |
18.83 |
44.736 |
7.477 |
T P |
| 1ihu_A |
540 |
257 |
158 |
2.08 |
18.99 |
43.965 |
7.237 |
T P |
| 3e70_C |
307 |
257 |
144 |
2.16 |
14.58 |
39.873 |
6.383 |
T P |
| 3b9q_A |
298 |
257 |
144 |
2.14 |
20.83 |
39.849 |
6.415 |
T P |
| 3dm5_A |
416 |
257 |
142 |
2.07 |
17.61 |
39.713 |
6.534 |
T P |
| 1fpm_A |
549 |
257 |
158 |
2.37 |
13.92 |
39.682 |
6.402 |
T P |
| 2og2_A |
302 |
257 |
144 |
2.13 |
19.44 |
39.539 |
6.446 |
T P |
| 3dm9_B |
305 |
257 |
144 |
2.12 |
14.58 |
39.502 |
6.486 |
T P |
| 2v3c_C |
403 |
257 |
140 |
2.10 |
17.86 |
39.132 |
6.358 |
T P |
| 1jpn_A |
296 |
257 |
142 |
2.12 |
17.61 |
39.127 |
6.400 |
T P |
| 1fts_A |
295 |
257 |
143 |
2.22 |
15.38 |
39.018 |
6.173 |
T P |
| 1qzx_A |
425 |
257 |
144 |
2.27 |
16.67 |
38.810 |
6.086 |
T P |
| 2ng1_A |
293 |
257 |
139 |
2.12 |
17.99 |
38.287 |
6.263 |
T P |
| 1ls1_A |
289 |
257 |
138 |
2.21 |
19.57 |
38.013 |
5.979 |
T P |
| 2ffh_A |
407 |
257 |
138 |
2.13 |
19.57 |
37.953 |
6.192 |
T P |
| 2j45_A |
297 |
257 |
138 |
2.18 |
18.84 |
37.830 |
6.054 |
T P |
| 1ffh_A |
287 |
257 |
133 |
2.10 |
20.30 |
37.781 |
6.049 |
T P |
| 1j8m_F |
295 |
257 |
143 |
2.29 |
16.08 |
37.750 |
5.973 |
T P |
| 2c03_B |
297 |
257 |
137 |
2.05 |
17.52 |
37.498 |
6.362 |
T P |
| 2j7p_A |
292 |
257 |
139 |
2.30 |
18.71 |
37.383 |
5.787 |
T P |
| 2j28_9 |
430 |
257 |
145 |
2.31 |
23.45 |
37.209 |
6.024 |
T P |
| 2j37_W |
479 |
257 |
131 |
2.02 |
20.61 |
37.204 |
6.183 |
T P |
| 1rj9_B |
282 |
257 |
137 |
2.25 |
18.25 |
37.049 |
5.836 |
T P |
| 1yrb_B |
260 |
257 |
142 |
2.39 |
16.20 |
36.759 |
5.708 |
T P |
| 2qy9_A |
300 |
257 |
135 |
2.13 |
14.81 |
36.745 |
6.057 |
T P |
| 1zu4_A |
305 |
257 |
133 |
2.25 |
19.55 |
36.317 |
5.668 |
T P |
| 1eg7_A |
549 |
257 |
153 |
2.58 |
15.03 |
36.267 |
5.699 |
T P |
| 1j8y_F |
291 |
257 |
133 |
2.09 |
15.79 |
36.100 |
6.074 |
T P |
| 1cr1_A |
245 |
257 |
99 |
2.29 |
15.15 |
28.144 |
4.142 |
T P |
| 1e0j_B |
288 |
257 |
104 |
2.35 |
17.31 |
27.835 |
4.246 |
T P |
| 3cr8_A |
493 |
257 |
102 |
2.27 |
18.63 |
27.472 |
4.307 |
T P |
| 2yvu_A |
179 |
257 |
96 |
2.14 |
16.67 |
26.934 |
4.282 |
T P |
| 2gks_B |
529 |
257 |
96 |
2.17 |
15.62 |
26.768 |
4.235 |
T P |
| 3e1s_A |
517 |
257 |
83 |
2.71 |
10.84 |
21.583 |
2.948 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

