LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_458.5wLII_11346_8
Total number of 3D structures: 46
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
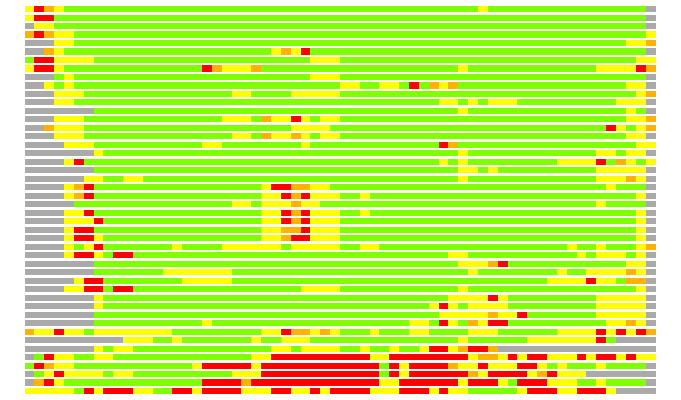
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 2b5a_A |
77 |
64 |
62 |
0.97 |
17.74 |
96.012 |
5.782 |
T P |
| 1y7y_A |
69 |
64 |
61 |
0.65 |
22.95 |
95.593 |
8.126 |
T P |
| 3f52_E |
78 |
64 |
62 |
0.97 |
22.58 |
95.230 |
5.800 |
T P |
| 1y9q_A |
178 |
64 |
63 |
1.40 |
15.87 |
94.716 |
4.199 |
T P |
| 3clc_B |
77 |
64 |
61 |
1.22 |
16.39 |
93.215 |
4.628 |
T P |
| 2ewt_A |
71 |
64 |
60 |
1.42 |
8.33 |
90.502 |
3.960 |
T P |
| 3f6w_A |
82 |
64 |
62 |
1.53 |
17.74 |
90.282 |
3.806 |
T P |
| 2ofy_A |
82 |
64 |
60 |
1.68 |
25.00 |
90.077 |
3.379 |
T P |
| 3eus_A |
85 |
64 |
60 |
1.23 |
15.00 |
89.021 |
4.502 |
T P |
| 3b7h_A |
76 |
64 |
60 |
1.72 |
21.67 |
87.932 |
3.293 |
T P |
| 1lmb_4 |
92 |
64 |
61 |
1.68 |
11.48 |
87.788 |
3.425 |
T P |
| 2bnm_A |
194 |
64 |
60 |
1.61 |
15.00 |
87.664 |
3.503 |
T P |
| 1adr_A |
76 |
64 |
56 |
1.01 |
14.29 |
87.116 |
5.066 |
T P |
| 1lli_B |
92 |
64 |
60 |
1.81 |
8.33 |
86.794 |
3.146 |
T P |
| 2ef8_A |
84 |
64 |
61 |
1.75 |
13.11 |
86.794 |
3.291 |
T P |
| 1rio_A |
97 |
64 |
60 |
1.82 |
10.00 |
86.239 |
3.132 |
T P |
| 3dnv_B |
71 |
64 |
59 |
1.61 |
22.03 |
85.958 |
3.445 |
T P |
| 1b0n_A |
103 |
64 |
56 |
1.17 |
16.07 |
85.273 |
4.395 |
T P |
| 2jvl_A |
107 |
64 |
58 |
1.65 |
15.52 |
85.060 |
3.315 |
T P |
| 2r1j_L |
66 |
64 |
56 |
1.32 |
14.29 |
84.635 |
3.932 |
T P |
| 1x57_A |
91 |
64 |
57 |
1.57 |
19.30 |
84.368 |
3.414 |
T P |
| 2axu_A |
303 |
64 |
56 |
1.66 |
17.86 |
84.272 |
3.174 |
T P |
| 2axz_A |
306 |
64 |
56 |
1.55 |
17.86 |
84.183 |
3.404 |
T P |
| 2qfc_A |
284 |
64 |
58 |
1.53 |
12.07 |
84.142 |
3.550 |
T P |
| 2grm_A |
315 |
64 |
56 |
1.57 |
17.86 |
84.135 |
3.346 |
T P |
| 2aw6_A |
287 |
64 |
56 |
1.64 |
17.86 |
83.512 |
3.227 |
T P |
| 2awi_A |
298 |
64 |
56 |
1.59 |
17.86 |
83.100 |
3.317 |
T P |
| 2axv_B |
303 |
64 |
55 |
1.49 |
18.18 |
83.073 |
3.457 |
T P |
| 3bdn_A |
234 |
64 |
59 |
1.98 |
11.86 |
82.538 |
2.831 |
T P |
| 1utx_A |
66 |
64 |
55 |
1.39 |
14.55 |
82.436 |
3.684 |
T P |
| 2cro_A |
65 |
64 |
55 |
1.39 |
21.82 |
82.168 |
3.702 |
T P |
| 2eby_A |
102 |
64 |
56 |
1.72 |
10.71 |
81.730 |
3.080 |
T P |
| 3cec_A |
91 |
64 |
55 |
1.65 |
7.27 |
81.572 |
3.144 |
T P |
| 3bs3_A |
62 |
64 |
55 |
1.44 |
14.55 |
80.585 |
3.561 |
T P |
| 1r69_A |
63 |
64 |
55 |
1.49 |
20.00 |
80.482 |
3.449 |
T P |
| 1r63_A |
63 |
64 |
55 |
1.62 |
20.00 |
79.886 |
3.190 |
T P |
| 1sq8_A |
64 |
64 |
55 |
1.76 |
20.00 |
79.537 |
2.961 |
T P |
| 2r63_A |
63 |
64 |
53 |
1.66 |
20.75 |
77.545 |
3.009 |
T P |
| 2iu7_A |
156 |
64 |
59 |
2.32 |
11.86 |
70.685 |
2.442 |
T P |
| 2ppx_A |
62 |
64 |
49 |
1.79 |
12.24 |
69.627 |
2.592 |
T P |
| 2k9q_A |
77 |
64 |
37 |
2.13 |
16.22 |
51.231 |
1.662 |
T P |
| 1a6d_B |
502 |
64 |
37 |
2.41 |
13.51 |
43.721 |
1.472 |
T P |
| 1b8p_A |
327 |
64 |
37 |
2.22 |
13.51 |
43.560 |
1.592 |
T P |
| 2ras_B |
204 |
64 |
31 |
2.45 |
16.13 |
41.050 |
1.215 |
T P |
| 3d5t_C |
323 |
64 |
32 |
2.13 |
12.50 |
40.709 |
1.435 |
T P |
| 1e0r_B |
154 |
64 |
33 |
2.74 |
0.00 |
33.858 |
1.162 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

