LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_483.5wLII_11346_127
Total number of 3D structures: 48
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
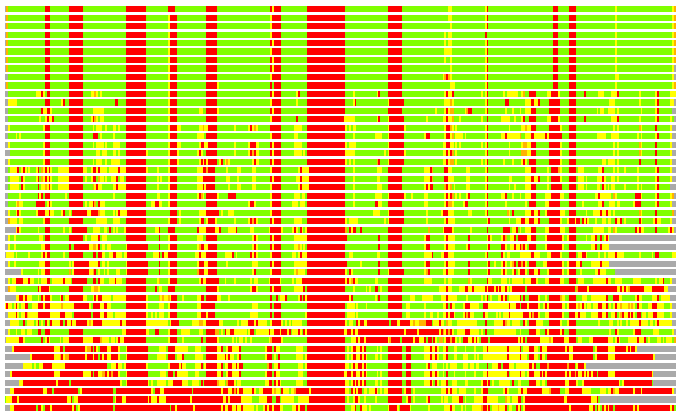
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 2g9a_A |
310 |
366 |
291 |
0.82 |
14.78 |
78.517 |
31.650 |
T P |
| 2h9m_A |
304 |
366 |
291 |
0.81 |
14.78 |
78.491 |
32.132 |
T P |
| 2gnq_A |
316 |
366 |
291 |
0.78 |
14.78 |
78.490 |
33.023 |
T P |
| 2h9l_A |
321 |
366 |
291 |
0.82 |
14.43 |
78.426 |
31.632 |
T P |
| 2h6n_B |
305 |
366 |
291 |
0.82 |
14.78 |
78.420 |
31.734 |
T P |
| 2g99_A |
304 |
366 |
291 |
0.81 |
14.78 |
78.419 |
31.938 |
T P |
| 2co0_A |
304 |
366 |
291 |
0.81 |
15.12 |
78.377 |
32.043 |
T P |
| 3emh_A |
300 |
366 |
290 |
0.80 |
14.83 |
78.219 |
32.382 |
T P |
| 2cnx_A |
306 |
366 |
288 |
0.79 |
15.28 |
77.942 |
32.201 |
T P |
| 2h14_A |
303 |
366 |
291 |
0.98 |
14.78 |
77.707 |
26.859 |
T P |
| 3frx_B |
313 |
366 |
278 |
1.46 |
12.59 |
71.526 |
17.827 |
T P |
| 1erj_C |
357 |
366 |
276 |
1.42 |
16.30 |
71.069 |
18.111 |
T P |
| 1vyh_C |
310 |
366 |
278 |
1.42 |
14.03 |
71.059 |
18.346 |
T P |
| 3dm0_A |
675 |
366 |
278 |
1.52 |
14.75 |
70.561 |
17.198 |
T P |
| 1got_B |
339 |
366 |
278 |
1.64 |
11.87 |
69.510 |
16.013 |
T P |
| 3fm0_A |
328 |
366 |
272 |
1.58 |
14.34 |
69.271 |
16.232 |
T P |
| 1tbg_A |
340 |
366 |
275 |
1.60 |
12.00 |
69.177 |
16.220 |
T P |
| 2bcj_B |
339 |
366 |
275 |
1.66 |
12.36 |
68.274 |
15.625 |
T P |
| 2pbi_D |
354 |
366 |
273 |
1.65 |
12.45 |
68.213 |
15.641 |
T P |
| 3cfs_B |
383 |
366 |
273 |
1.83 |
11.36 |
67.054 |
14.127 |
T P |
| 3cfv_B |
393 |
366 |
273 |
1.83 |
10.99 |
67.048 |
14.175 |
T P |
| 3c99_A |
380 |
366 |
273 |
1.85 |
11.36 |
66.720 |
14.024 |
T P |
| 2hes_X |
308 |
366 |
268 |
1.72 |
11.57 |
66.716 |
14.721 |
T P |
| 1p22_A |
402 |
366 |
262 |
1.61 |
9.54 |
65.749 |
15.315 |
T P |
| 1gxr_A |
335 |
366 |
266 |
1.83 |
8.27 |
65.289 |
13.762 |
T P |
| 1a0r_B |
339 |
366 |
269 |
1.80 |
11.90 |
61.925 |
14.193 |
T P |
| 1nr0_A |
610 |
366 |
254 |
1.78 |
9.84 |
59.004 |
13.498 |
T P |
| 2pm7_D |
288 |
366 |
227 |
1.69 |
10.57 |
55.091 |
12.700 |
T P |
| 2pm6_D |
288 |
366 |
228 |
1.78 |
10.09 |
54.061 |
12.150 |
T P |
| 3ei4_B |
368 |
366 |
265 |
1.98 |
12.45 |
53.935 |
12.730 |
T P |
| 2pm9_B |
280 |
366 |
228 |
1.86 |
10.09 |
53.151 |
11.629 |
T P |
| 3bg1_A |
285 |
366 |
226 |
1.78 |
11.06 |
52.585 |
12.038 |
T P |
| 2pm9_A |
384 |
366 |
264 |
2.00 |
10.98 |
52.187 |
12.552 |
T P |
| 1nex_B |
444 |
366 |
205 |
1.94 |
12.68 |
49.633 |
10.038 |
T P |
| 1pi6_A |
606 |
366 |
251 |
2.12 |
12.75 |
49.531 |
11.290 |
T P |
| 2b4e_A |
386 |
366 |
243 |
2.22 |
10.70 |
49.295 |
10.471 |
T P |
| 2aq5_A |
395 |
366 |
244 |
2.25 |
10.25 |
49.049 |
10.374 |
T P |
| 2zkq_a |
306 |
366 |
243 |
2.54 |
13.17 |
46.205 |
9.193 |
T P |
| 2ovr_B |
442 |
366 |
221 |
2.10 |
12.67 |
45.763 |
10.032 |
T P |
| 1r5m_A |
351 |
366 |
209 |
2.35 |
6.70 |
38.386 |
8.516 |
T P |
| 2w8b_B |
409 |
366 |
184 |
2.35 |
12.50 |
34.354 |
7.502 |
T P |
| 1crz_A |
403 |
366 |
183 |
2.35 |
12.02 |
34.125 |
7.481 |
T P |
| 2ojh_A |
277 |
366 |
179 |
2.17 |
14.53 |
34.088 |
7.869 |
T P |
| 2hqs_A |
412 |
366 |
186 |
2.36 |
12.90 |
33.862 |
7.563 |
T P |
| 1c5k_A |
397 |
366 |
169 |
2.26 |
14.79 |
32.004 |
7.149 |
T P |
| 2dcm_A |
662 |
366 |
150 |
2.46 |
10.67 |
27.006 |
5.869 |
T P |
| 2z3z_A |
651 |
366 |
143 |
2.44 |
11.19 |
26.683 |
5.632 |
T P |
| 2d5l_A |
665 |
366 |
138 |
2.49 |
13.04 |
25.088 |
5.322 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

