LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_505.5wLII_11364_46
Total number of 3D structures: 38
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
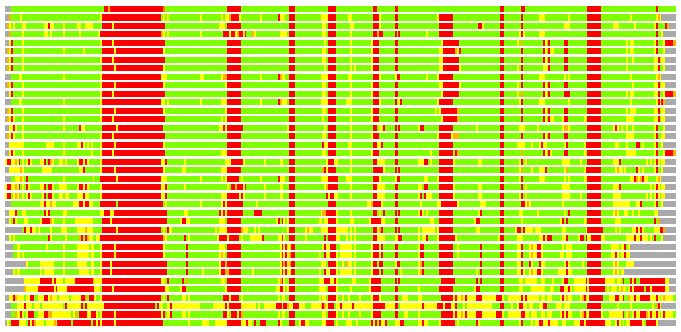
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 1erj_C |
357 |
344 |
282 |
0.61 |
18.09 |
82.005 |
39.658 |
T P |
| 1got_B |
339 |
344 |
271 |
1.37 |
11.81 |
74.739 |
18.465 |
T P |
| 1vyh_C |
310 |
344 |
268 |
1.34 |
8.58 |
74.100 |
18.548 |
T P |
| 2pbi_D |
354 |
344 |
266 |
1.37 |
10.90 |
73.949 |
18.040 |
T P |
| 2h6n_B |
305 |
344 |
265 |
1.23 |
11.70 |
73.916 |
19.992 |
T P |
| 2gnq_A |
316 |
344 |
266 |
1.25 |
11.65 |
73.747 |
19.667 |
T P |
| 2h9m_A |
304 |
344 |
264 |
1.20 |
12.12 |
73.698 |
20.265 |
T P |
| 2h9l_A |
321 |
344 |
266 |
1.28 |
12.03 |
73.638 |
19.345 |
T P |
| 1tbg_A |
340 |
344 |
267 |
1.41 |
11.99 |
73.532 |
17.712 |
T P |
| 2co0_A |
304 |
344 |
265 |
1.27 |
12.08 |
73.529 |
19.402 |
T P |
| 2g99_A |
304 |
344 |
264 |
1.25 |
11.74 |
73.508 |
19.528 |
T P |
| 2bcj_B |
339 |
344 |
267 |
1.39 |
11.99 |
73.476 |
17.927 |
T P |
| 2cnx_A |
306 |
344 |
265 |
1.27 |
12.08 |
73.392 |
19.367 |
T P |
| 2g9a_A |
310 |
344 |
265 |
1.30 |
11.70 |
73.127 |
18.872 |
T P |
| 3fm0_A |
328 |
344 |
264 |
1.36 |
10.61 |
72.919 |
18.041 |
T P |
| 3emh_A |
300 |
344 |
266 |
1.36 |
12.03 |
72.722 |
18.247 |
T P |
| 3dm0_A |
675 |
344 |
263 |
1.43 |
12.17 |
72.254 |
17.190 |
T P |
| 2h14_A |
303 |
344 |
265 |
1.42 |
11.32 |
72.139 |
17.436 |
T P |
| 3cfs_B |
383 |
344 |
262 |
1.63 |
7.25 |
71.299 |
15.171 |
T P |
| 3frx_B |
313 |
344 |
263 |
1.56 |
11.03 |
71.253 |
15.882 |
T P |
| 1a0r_B |
339 |
344 |
264 |
1.64 |
11.74 |
71.215 |
15.189 |
T P |
| 3cfv_B |
393 |
344 |
259 |
1.57 |
7.34 |
71.114 |
15.467 |
T P |
| 3c99_A |
380 |
344 |
264 |
1.70 |
7.58 |
70.674 |
14.703 |
T P |
| 1gxr_A |
335 |
344 |
256 |
1.60 |
10.16 |
69.298 |
15.045 |
T P |
| 2hes_X |
308 |
344 |
254 |
1.51 |
8.66 |
69.108 |
15.815 |
T P |
| 1p22_A |
402 |
344 |
251 |
1.47 |
8.76 |
68.815 |
15.999 |
T P |
| 1nr0_A |
610 |
344 |
255 |
1.87 |
12.94 |
65.876 |
12.944 |
T P |
| 3ei4_B |
368 |
344 |
250 |
1.79 |
11.60 |
65.473 |
13.193 |
T P |
| 2pm7_D |
288 |
344 |
242 |
1.74 |
7.44 |
64.470 |
13.177 |
T P |
| 2pm6_D |
288 |
344 |
241 |
1.75 |
8.30 |
64.338 |
13.053 |
T P |
| 3bg1_A |
285 |
344 |
234 |
1.73 |
7.26 |
63.127 |
12.789 |
T P |
| 2pm9_B |
280 |
344 |
237 |
1.81 |
6.75 |
60.166 |
12.425 |
T P |
| 1nex_B |
444 |
344 |
223 |
1.90 |
10.31 |
56.518 |
11.157 |
T P |
| 2ovr_B |
442 |
344 |
214 |
1.85 |
9.35 |
55.481 |
10.979 |
T P |
| 2aq5_A |
395 |
344 |
247 |
2.26 |
8.50 |
54.218 |
10.484 |
T P |
| 2zkq_a |
306 |
344 |
251 |
2.45 |
10.36 |
53.700 |
9.843 |
T P |
| 2b4e_A |
386 |
344 |
245 |
2.24 |
8.57 |
52.139 |
10.478 |
T P |
| 1r5m_A |
351 |
344 |
212 |
2.15 |
10.38 |
45.446 |
9.420 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

