LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_51.5wLII_10933_47
Total number of 3D structures: 11
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
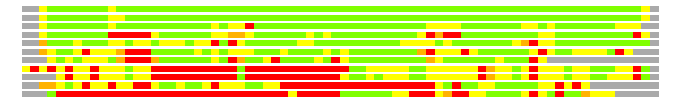
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 2nsm_A |
390 |
74 |
71 |
0.66 |
16.90 |
95.620 |
9.353 |
T P |
| 1h8l_A |
380 |
74 |
71 |
0.90 |
9.86 |
94.867 |
7.107 |
T P |
| 1uwy_A |
393 |
74 |
69 |
1.75 |
18.84 |
85.254 |
3.735 |
T P |
| 1nkg_A |
508 |
74 |
62 |
1.91 |
8.06 |
77.144 |
3.092 |
T P |
| 3g3l_A |
291 |
74 |
68 |
1.93 |
16.18 |
76.526 |
3.348 |
T P |
| 1qho_A |
686 |
74 |
61 |
2.15 |
4.92 |
65.089 |
2.713 |
T P |
| 3cmg_A |
661 |
74 |
51 |
1.96 |
17.65 |
56.146 |
2.472 |
T P |
| 1xqm_A |
227 |
74 |
44 |
2.60 |
13.64 |
40.008 |
1.627 |
T P |
| 2a50_B |
167 |
74 |
43 |
2.63 |
13.95 |
39.059 |
1.576 |
T P |
| 1xmz_B |
231 |
74 |
38 |
2.54 |
2.63 |
36.576 |
1.439 |
T P |
| 1xi9_C |
393 |
74 |
27 |
2.46 |
11.11 |
26.989 |
1.057 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

