LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_516.5wLII_11389_17
Total number of 3D structures: 106
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
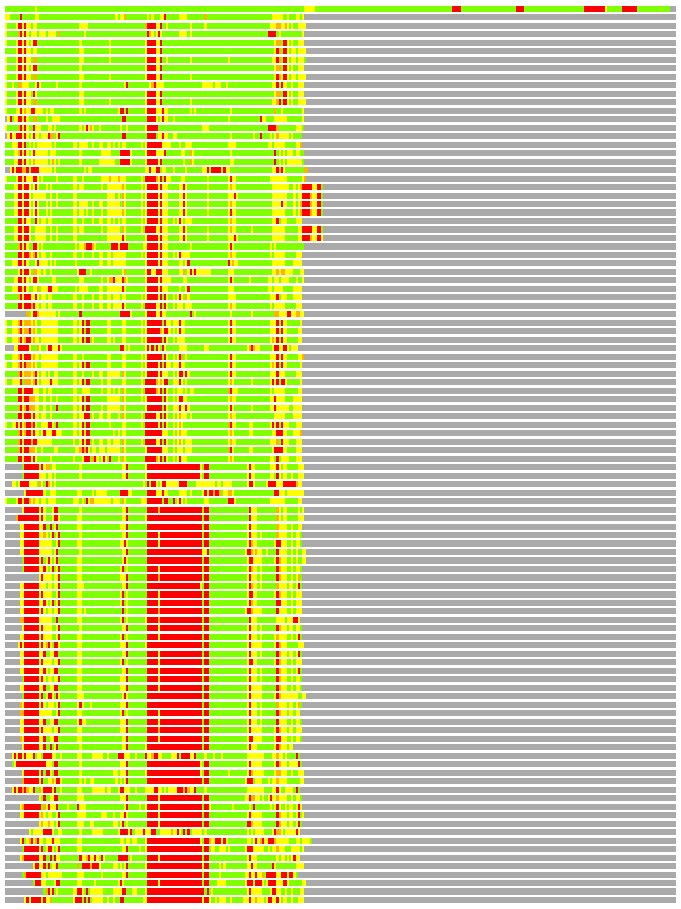
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 2hyx_A |
333 |
316 |
289 |
0.70 |
33.56 |
90.712 |
36.245 |
T P |
| 3eyt_C |
157 |
316 |
139 |
1.54 |
24.46 |
40.728 |
8.450 |
T P |
| 3c71_A |
140 |
316 |
134 |
1.75 |
21.64 |
39.239 |
7.260 |
T P |
| 2ywi_A |
190 |
316 |
133 |
1.67 |
15.04 |
39.202 |
7.510 |
T P |
| 2f9s_A |
138 |
316 |
129 |
1.43 |
21.71 |
38.971 |
8.413 |
T P |
| 2h19_A |
138 |
316 |
131 |
1.57 |
21.37 |
38.892 |
7.839 |
T P |
| 2h1a_B |
139 |
316 |
131 |
1.56 |
21.37 |
38.859 |
7.895 |
T P |
| 1st9_A |
137 |
316 |
129 |
1.45 |
21.71 |
38.796 |
8.344 |
T P |
| 3c73_B |
138 |
316 |
131 |
1.60 |
22.14 |
38.760 |
7.723 |
T P |
| 2b5x_A |
148 |
316 |
136 |
1.82 |
26.47 |
38.752 |
7.100 |
T P |
| 2h1b_A |
138 |
316 |
130 |
1.59 |
21.54 |
38.730 |
7.685 |
T P |
| 2h1g_B |
137 |
316 |
131 |
1.60 |
20.61 |
38.515 |
7.685 |
T P |
| 1kng_A |
144 |
316 |
130 |
1.69 |
24.62 |
37.571 |
7.259 |
T P |
| 1zzo_A |
134 |
316 |
128 |
1.70 |
14.06 |
37.299 |
7.124 |
T P |
| 3fkf_A |
141 |
316 |
128 |
1.70 |
19.53 |
36.912 |
7.105 |
T P |
| 1lu4_A |
134 |
316 |
123 |
1.75 |
16.26 |
36.229 |
6.653 |
T P |
| 1xcc_A |
219 |
316 |
130 |
2.00 |
10.00 |
35.621 |
6.188 |
T P |
| 2b1k_A |
149 |
316 |
127 |
1.90 |
16.54 |
35.599 |
6.352 |
T P |
| 2g0f_A |
149 |
316 |
126 |
1.86 |
16.67 |
35.488 |
6.412 |
T P |
| 2p5q_A |
161 |
316 |
120 |
1.79 |
18.33 |
35.409 |
6.351 |
T P |
| 1xxu_A |
153 |
316 |
129 |
1.90 |
16.28 |
35.402 |
6.456 |
T P |
| 2nvl_A |
242 |
316 |
131 |
2.00 |
12.21 |
35.236 |
6.240 |
T P |
| 2e2g_B |
239 |
316 |
132 |
2.05 |
12.88 |
34.894 |
6.153 |
T P |
| 2cv4_D |
242 |
316 |
131 |
2.04 |
12.21 |
34.894 |
6.113 |
T P |
| 1vgs_D |
242 |
316 |
131 |
2.03 |
12.98 |
34.894 |
6.147 |
T P |
| 1qmv_A |
196 |
316 |
128 |
1.97 |
13.28 |
34.824 |
6.193 |
T P |
| 1x0r_A |
244 |
316 |
132 |
2.04 |
12.88 |
34.563 |
6.155 |
T P |
| 2zct_D |
238 |
316 |
131 |
2.03 |
12.21 |
34.416 |
6.146 |
T P |
| 2ywn_A |
150 |
316 |
120 |
1.70 |
18.33 |
34.343 |
6.682 |
T P |
| 1uul_A |
194 |
316 |
128 |
2.04 |
10.16 |
34.342 |
5.986 |
T P |
| 1prx_A |
220 |
316 |
128 |
1.96 |
10.94 |
34.087 |
6.224 |
T P |
| 1jfu_B |
177 |
316 |
128 |
2.13 |
17.97 |
33.659 |
5.737 |
T P |
| 2cx4_A |
161 |
316 |
127 |
1.93 |
13.39 |
33.037 |
6.255 |
T P |
| 2v2g_A |
220 |
316 |
127 |
1.99 |
11.02 |
32.911 |
6.086 |
T P |
| 2pn8_A |
191 |
316 |
126 |
1.98 |
15.08 |
32.858 |
6.054 |
T P |
| 2z9s_A |
197 |
316 |
126 |
1.97 |
15.08 |
32.779 |
6.078 |
T P |
| 1z5y_E |
136 |
316 |
115 |
1.96 |
18.26 |
32.635 |
5.584 |
T P |
| 1yf0_A |
167 |
316 |
124 |
2.02 |
19.35 |
32.433 |
5.845 |
T P |
| 1yf1_A |
166 |
316 |
123 |
2.01 |
19.51 |
32.401 |
5.839 |
T P |
| 3emp_B |
162 |
316 |
123 |
2.00 |
18.70 |
32.195 |
5.868 |
T P |
| 3erw_A |
134 |
316 |
115 |
1.93 |
19.13 |
32.001 |
5.669 |
T P |
| 1n8j_A |
186 |
316 |
125 |
2.00 |
17.60 |
31.932 |
5.948 |
T P |
| 1yep_A |
168 |
316 |
124 |
2.03 |
19.35 |
31.446 |
5.829 |
T P |
| 2i81_A |
193 |
316 |
127 |
2.09 |
14.17 |
31.402 |
5.793 |
T P |
| 1yex_A |
167 |
316 |
122 |
2.09 |
18.85 |
31.039 |
5.572 |
T P |
| 1zye_A |
162 |
316 |
126 |
2.03 |
11.11 |
30.905 |
5.907 |
T P |
| 2h01_A |
174 |
316 |
122 |
2.06 |
17.21 |
30.141 |
5.660 |
T P |
| 2feg_A |
169 |
316 |
122 |
2.09 |
16.39 |
30.071 |
5.582 |
T P |
| 1e2y_G |
173 |
316 |
124 |
2.01 |
12.10 |
29.637 |
5.869 |
T P |
| 1we0_A |
166 |
316 |
117 |
1.97 |
15.38 |
29.490 |
5.666 |
T P |
| 1zof_B |
175 |
316 |
119 |
2.10 |
14.29 |
29.439 |
5.397 |
T P |
| 2rii_B |
185 |
316 |
123 |
2.10 |
16.26 |
29.416 |
5.599 |
T P |
| 2c0d_B |
175 |
316 |
123 |
2.16 |
10.57 |
29.263 |
5.442 |
T P |
| 1qq2_A |
173 |
316 |
119 |
1.99 |
17.65 |
29.234 |
5.691 |
T P |
| 1xwa_A |
111 |
316 |
95 |
1.55 |
16.84 |
28.159 |
5.749 |
T P |
| 1xwb_A |
106 |
316 |
96 |
1.53 |
16.67 |
28.006 |
5.898 |
T P |
| 1o73_A |
144 |
316 |
111 |
2.17 |
13.51 |
27.974 |
4.889 |
T P |
| 2b1l_B |
129 |
316 |
108 |
2.13 |
18.52 |
27.947 |
4.845 |
T P |
| 2bmx_B |
178 |
316 |
121 |
2.25 |
16.53 |
27.912 |
5.157 |
T P |
| 2yzu_A |
104 |
316 |
94 |
1.62 |
23.40 |
27.694 |
5.470 |
T P |
| 2cvk_A |
105 |
316 |
93 |
1.59 |
23.66 |
27.694 |
5.512 |
T P |
| 2i4a_A |
107 |
316 |
93 |
1.80 |
20.43 |
27.225 |
4.887 |
T P |
| 2fch_D |
107 |
316 |
95 |
1.87 |
20.00 |
27.147 |
4.813 |
T P |
| 2h74_A |
108 |
316 |
94 |
1.90 |
19.15 |
27.004 |
4.696 |
T P |
| 1t00_A |
112 |
316 |
96 |
1.97 |
19.79 |
26.955 |
4.628 |
T P |
| 1nw2_A |
105 |
316 |
92 |
1.77 |
25.00 |
26.892 |
4.929 |
T P |
| 2h72_A |
107 |
316 |
92 |
1.81 |
20.65 |
26.850 |
4.828 |
T P |
| 2trx_A |
108 |
316 |
92 |
1.85 |
19.57 |
26.787 |
4.726 |
T P |
| 2eio_A |
107 |
316 |
95 |
1.99 |
18.95 |
26.738 |
4.556 |
T P |
| 2h71_A |
107 |
316 |
93 |
1.91 |
20.43 |
26.711 |
4.634 |
T P |
| 2h76_A |
108 |
316 |
92 |
1.86 |
21.74 |
26.663 |
4.693 |
T P |
| 3dyr_A |
111 |
316 |
93 |
1.89 |
19.35 |
26.660 |
4.670 |
T P |
| 2h6z_A |
108 |
316 |
93 |
1.88 |
18.28 |
26.654 |
4.694 |
T P |
| 2eir_A |
107 |
316 |
93 |
1.90 |
19.35 |
26.649 |
4.659 |
T P |
| 2h73_A |
108 |
316 |
94 |
1.95 |
19.15 |
26.648 |
4.593 |
T P |
| 2o7k_A |
103 |
316 |
92 |
1.83 |
18.48 |
26.632 |
4.778 |
T P |
| 3dxb_D |
216 |
316 |
92 |
1.85 |
19.57 |
26.601 |
4.710 |
T P |
| 2h6y_A |
107 |
316 |
93 |
1.94 |
19.35 |
26.581 |
4.562 |
T P |
| 2fd3_A |
108 |
316 |
92 |
1.87 |
19.57 |
26.504 |
4.680 |
T P |
| 1zzy_A |
106 |
316 |
93 |
1.88 |
19.35 |
26.498 |
4.691 |
T P |
| 2tir_A |
108 |
316 |
93 |
1.93 |
19.35 |
26.472 |
4.586 |
T P |
| 1nsw_A |
105 |
316 |
92 |
1.86 |
23.91 |
26.395 |
4.704 |
T P |
| 1zcp_A |
106 |
316 |
93 |
1.96 |
18.28 |
26.382 |
4.521 |
T P |
| 2h70_A |
107 |
316 |
94 |
1.98 |
19.15 |
26.360 |
4.516 |
T P |
| 1tho_A |
109 |
316 |
92 |
1.88 |
19.57 |
26.350 |
4.653 |
T P |
| 2o8v_B |
108 |
316 |
94 |
1.98 |
19.15 |
26.336 |
4.527 |
T P |
| 1txx_A |
108 |
316 |
91 |
1.86 |
20.88 |
26.141 |
4.641 |
T P |
| 2i1u_A |
108 |
316 |
87 |
1.61 |
24.14 |
26.110 |
5.101 |
T P |
| 2h30_A |
151 |
316 |
114 |
2.17 |
19.30 |
26.090 |
5.021 |
T P |
| 2eiq_A |
107 |
316 |
90 |
1.81 |
20.00 |
26.080 |
4.718 |
T P |
| 2gzy_A |
104 |
316 |
92 |
1.96 |
20.65 |
25.802 |
4.458 |
T P |
| 1quw_A |
105 |
316 |
91 |
1.97 |
24.18 |
25.721 |
4.386 |
T P |
| 2fy6_A |
143 |
316 |
115 |
2.25 |
20.00 |
25.557 |
4.895 |
T P |
| 2h75_B |
108 |
316 |
91 |
1.98 |
18.68 |
25.539 |
4.381 |
T P |
| 1srx_A |
108 |
316 |
91 |
1.95 |
17.58 |
25.459 |
4.430 |
T P |
| 1keb_A |
108 |
316 |
95 |
2.09 |
17.89 |
25.458 |
4.343 |
T P |
| 1f6m_C |
108 |
316 |
91 |
2.06 |
17.58 |
25.175 |
4.220 |
T P |
| 2jzr_A |
144 |
316 |
111 |
2.28 |
19.82 |
24.626 |
4.665 |
T P |
| 3f8u_A |
469 |
316 |
98 |
2.19 |
14.29 |
23.970 |
4.277 |
T P |
| 1rqm_A |
105 |
316 |
90 |
2.07 |
25.56 |
23.743 |
4.154 |
T P |
| 1oaz_A |
115 |
316 |
84 |
1.97 |
14.29 |
23.364 |
4.063 |
T P |
| 1gl8_A |
104 |
316 |
85 |
1.91 |
18.82 |
22.922 |
4.228 |
T P |
| 2alb_A |
113 |
316 |
82 |
2.01 |
13.41 |
22.612 |
3.879 |
T P |
| 1mek_A |
120 |
316 |
84 |
1.96 |
13.10 |
21.975 |
4.083 |
T P |
| 2i9h_A |
103 |
316 |
85 |
2.11 |
21.18 |
20.374 |
3.839 |
T P |
| 1wmj_A |
122 |
316 |
86 |
2.13 |
15.12 |
20.099 |
3.858 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

