LGA
Sequence Independent Analysis (LGA)
Frame of reference: Cat.Q2246_545_547.5wLII_11391_11
Total number of 3D structures: 45
LGA calculations using distance cutoff DIST: 4.0 A
Residues superimposed below 2.00 A: GREEN
Residues superimposed below 4.00 A: YELLOW
Residues superimposed below 6.00 A: ORANGE
Residues superimposed below 8.00 A: BROWN
Residues superimposed above 8.00 A or not aligned: RED
Terminal residues not aligned: GREY
Structure Deviation Summary
Calculations based on one final LGA superposition
(Bar representation of 3D plots, TEXT)
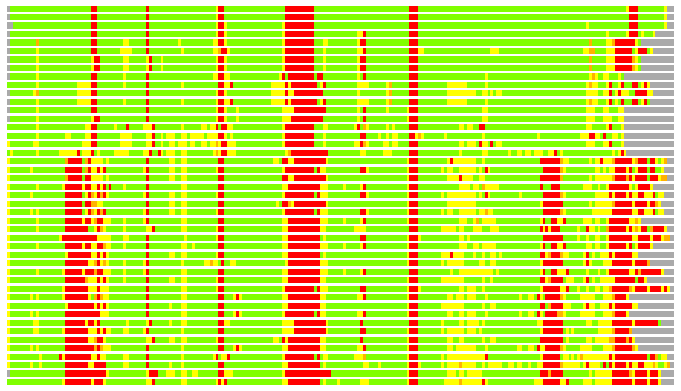
Structures ordered by LGA_S - score
| Structure |
NS |
NT |
N(dist=4.0) |
RMSD(N) |
Seq_ID(N) |
LGA_S |
LGA_Q |
PLOTS |
| 1g0u_K |
212 |
230 |
206 |
0.48 |
18.93 |
89.436 |
35.561 |
T P |
| 1ryp_L |
212 |
230 |
206 |
0.51 |
18.93 |
89.415 |
33.761 |
T P |
| 1g65_K |
211 |
230 |
205 |
0.90 |
18.54 |
87.586 |
20.529 |
T P |
| 1iru_L |
201 |
230 |
201 |
0.84 |
17.91 |
86.034 |
21.303 |
T P |
| 1yar_H |
203 |
230 |
191 |
1.32 |
11.52 |
79.115 |
13.483 |
T P |
| 1j2q_H |
202 |
230 |
194 |
1.46 |
16.49 |
78.991 |
12.408 |
T P |
| 3c91_H |
203 |
230 |
192 |
1.36 |
11.46 |
78.974 |
13.158 |
T P |
| 1pma_B |
203 |
230 |
192 |
1.36 |
11.46 |
78.947 |
13.158 |
T P |
| 1iru_I |
220 |
230 |
191 |
1.27 |
15.71 |
78.814 |
13.970 |
T P |
| 1ryp_H |
205 |
230 |
197 |
1.55 |
17.77 |
78.772 |
11.963 |
T P |
| 1iru_H |
202 |
230 |
197 |
1.60 |
19.29 |
78.545 |
11.568 |
T P |
| 1g65_N |
196 |
230 |
196 |
1.56 |
18.37 |
78.244 |
11.837 |
T P |
| 1ryp_I |
222 |
230 |
192 |
1.38 |
15.62 |
77.988 |
13.017 |
T P |
| 3bdm_H |
222 |
230 |
192 |
1.37 |
15.62 |
77.973 |
13.068 |
T P |
| 1ryp_M |
222 |
230 |
189 |
1.62 |
15.87 |
75.360 |
10.988 |
T P |
| 1iru_J |
204 |
230 |
191 |
1.78 |
10.99 |
74.336 |
10.147 |
T P |
| 1iru_M |
213 |
230 |
191 |
1.79 |
16.23 |
74.088 |
10.082 |
T P |
| 1iru_K |
199 |
230 |
190 |
1.83 |
15.79 |
73.696 |
9.863 |
T P |
| 1ryp_E |
242 |
230 |
183 |
1.85 |
13.11 |
73.198 |
9.389 |
T P |
| 3bdm_C |
241 |
230 |
182 |
1.71 |
13.19 |
72.977 |
10.051 |
T P |
| 1g0u_D |
230 |
230 |
182 |
1.83 |
12.64 |
72.774 |
9.449 |
T P |
| 3bdm_F |
244 |
230 |
179 |
1.65 |
15.08 |
72.233 |
10.248 |
T P |
| 1ryp_D |
241 |
230 |
183 |
1.81 |
13.11 |
72.053 |
9.557 |
T P |
| 3bdm_D |
242 |
230 |
180 |
1.77 |
12.78 |
71.995 |
9.604 |
T P |
| 1g0u_C |
238 |
230 |
183 |
1.80 |
12.57 |
71.956 |
9.627 |
T P |
| 1g0u_F |
242 |
230 |
180 |
1.74 |
13.89 |
71.382 |
9.798 |
T P |
| 1iru_G |
245 |
230 |
183 |
1.78 |
12.57 |
71.335 |
9.741 |
T P |
| 1j2q_A |
237 |
230 |
176 |
1.65 |
13.64 |
71.224 |
10.083 |
T P |
| 1ryp_G |
244 |
230 |
180 |
1.72 |
13.89 |
71.078 |
9.912 |
T P |
| 3bdm_B |
244 |
230 |
177 |
1.75 |
12.99 |
70.969 |
9.573 |
T P |
| 1ryp_B |
250 |
230 |
178 |
1.70 |
14.04 |
70.860 |
9.905 |
T P |
| 1z7q_G |
243 |
230 |
179 |
1.78 |
13.97 |
70.737 |
9.534 |
T P |
| 1g0u_B |
235 |
230 |
179 |
1.76 |
13.41 |
70.591 |
9.637 |
T P |
| 1j2p_A |
243 |
230 |
179 |
1.79 |
13.41 |
70.549 |
9.477 |
T P |
| 1ya7_A |
227 |
230 |
177 |
1.73 |
13.56 |
70.387 |
9.646 |
T P |
| 1ryp_C |
244 |
230 |
176 |
1.72 |
13.64 |
70.308 |
9.662 |
T P |
| 1yar_A |
221 |
230 |
175 |
1.69 |
13.71 |
70.235 |
9.759 |
T P |
| 1iru_F |
238 |
230 |
176 |
1.71 |
11.36 |
70.039 |
9.742 |
T P |
| 1iru_B |
233 |
230 |
175 |
1.73 |
14.86 |
69.876 |
9.544 |
T P |
| 1iru_E |
234 |
230 |
175 |
1.73 |
12.57 |
69.853 |
9.584 |
T P |
| 1yau_A |
222 |
230 |
176 |
1.74 |
13.64 |
69.836 |
9.557 |
T P |
| 1iru_D |
243 |
230 |
179 |
1.89 |
10.06 |
67.988 |
8.982 |
T P |
| 1iru_C |
250 |
230 |
184 |
1.98 |
11.96 |
67.673 |
8.859 |
T P |
| 2z5c_C |
189 |
230 |
167 |
1.71 |
11.98 |
67.067 |
9.234 |
T P |
| 1iru_A |
244 |
230 |
175 |
1.94 |
7.43 |
65.025 |
8.560 |
T P |
NS : Total number of residues in Structure (rotated structure)
NT : Total number of residues in TARGET (frame of reference)
N : Total number of residues superimposed under 4.0 Angstrom distance cutoff
RMSD : RMS deviation calculated on all N residues superimposed under 4.0 Angstrom distance cutoff
Seq_Id : Sequence Identity. Percent of identical residues from the total of N aligned.
LGA_S : Structure similarity score calculated by internal LGA procedure (see LGA paper for details)
LGA_Q : Score (how tight is the superposition) calculated by the formula: Q = 0.1*N/(0.1+RMSD)
PLOTS : T - Flat text file (output from LGA program, rotated structure)
PLOTS : P - Plot of superimposed structures (3D plot colored as bars)
Citing LGA:
Zemla A., "LGA - a Method for Finding 3D Similarities in Protein Structures",
Nucleic Acids Research, 2003, Vol. 31, No. 13, pp. 3370-3374.
[MEDLINE]

