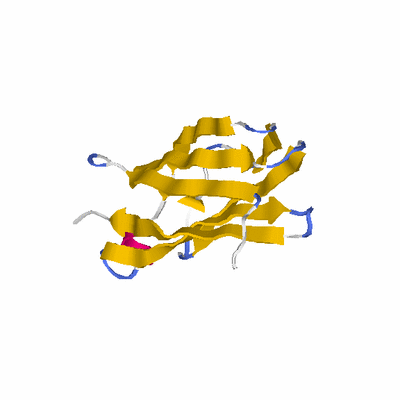
##########
# PROTEIN: Q2246_545_120.5wLII_11077_13 442 120 # Category_C2
# Length: 442
# Weight: 49823.43
# Similarity: UniProt_B6ARC1, BLAST_B6ARC1, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 22.17 M_00 2r39_A 118 1 4e-22 17.000 98:113 (336-438:5-115)
# Evl 45.70 M_00 1y4z_B 512 1 2e-47 14.000 202:248 (196-437:173-380)
# Sid 3.17 M_32 1dur_A 55 1 0.073 35.000 14:14 (243-256:36-49)
# Cov 49.77 M_00 1y4z_B 512 1 1e-29 10.000 220:250 (185-411:163-405)
# LaL 13.57 M_08 1jb0_C 80 1 2e-07 18.000 60:60 (204-263:8-67)
# OvN 45.02 M_26 z0gz 522 0 4e-05 7.000 199:225 (99-298:22-245)
# OvC 46.15 M_02 z0gz 522 0 2e-46 11.000 204:253 (196-442:171-382)
PDB: 2r39 PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 29-AUG-07 2R39
TITLE CRYSTAL STRUCTURE OF FIXG-RELATED PROTEIN FROM VIBRIO
TITLE 2 PARAHAEMOLYTICUS
SOURCE 2 ORGANISM_SCIENTIFIC: VIBRIO PARAHAEMOLYTICUS RIMD 2210633;
SOURCE 3 ORGANISM_TAXID: 223926;
KEYWDS FIXG-RELATED PROTEIN, VIBRIO PARAHAEMOLYTICUS, STRUCTURAL
KEYWDS 2 GENOMICS, PSI-2, PROTEIN STRUCTURE INITIATIVE, MIDWEST
KEYWDS 3 CENTER FOR STRUCTURAL GENOMICS, MCSG, 4FE-4S, IRON, IRON-
KEYWDS 4 SULFUR, METAL-BINDING, UNKNOWN FUNCTION
SCOP: none
PDB: 1y4z PDBsum
HEADER OXIDOREDUCTASE 01-DEC-04 1Y4Z
TITLE THE CRYSTAL STRUCTURE OF NITRATE REDUCTASE A, NARGHI, IN
TITLE 2 COMPLEX WITH THE Q-SITE INHIBITOR PENTACHLOROPHENOL
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
SOURCE 11 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
COMPND 6 EC: 1.7.99.4;
COMPND 13 EC: 1.7.99.4;
COMPND 21 EC: 1.7.99.4;
KEYWDS NITRATE REDUCTION, ELECTRON TRANSFER, MEMBRANE PROTEIN, Q-
KEYWDS 2 SITE, OXIDOREDUCTASE
SCOP: b.52 Double psi beta-barrel
SCOP: b.52.2 ADC-like
SCOP: b.52.2.2 Formate dehydrogenase/DMSO reductase, C-terminal domain
SCOP: c.81 Formate dehydrogenase/DMSO reductase, domains 1-3
SCOP: c.81.1 Formate dehydrogenase/DMSO reductase, domains 1-3
SCOP: c.81.1.1 Formate dehydrogenase/DMSO reductase, domains 1-3
SCOP: f.21 Heme-binding four-helical bundle
SCOP: f.21.3 Respiratory nitrate reductase 1 gamma chain
SCOP: f.21.3.1 Respiratory nitrate reductase 1 gamma chain
PDB: 1dur PDBsum
HEADER ELECTRON TRANSPORT 18-JAN-00 1DUR
TITLE REPLACEMENT FOR 1FDX 2(4FE4S) FERREDOXIN FROM (NOW)
TITLE 2 PEPTOSTREPTOCOCCUS ASACCHAROLYTICUS
SOURCE 2 ORGANISM_SCIENTIFIC: PEPTONIPHILUS ASACCHAROLYTICUS;
SOURCE 3 ORGANISM_TAXID: 1258;
KEYWDS TWO 4FE-4S CLUSTERS, ELECTRON TRANSPORT
SCOP: d.58 Ferredoxin-like
SCOP: d.58.1 4Fe-4S ferredoxins
SCOP: d.58.1.1 Short-chain ferredoxins
PDB: 1jb0 PDBsum
HEADER PHOTOSYNTHESIS 01-JUN-01 1JB0
TITLE CRYSTAL STRUCTURE OF PHOTOSYSTEM I: A PHOTOSYNTHETIC
TITLE 2 REACTION CENTER AND CORE ANTENNA SYSTEM FROM CYANOBACTERIA
SOURCE 2 ORGANISM_SCIENTIFIC: SYNECHOCOCCUS ELONGATUS;
SOURCE 3 ORGANISM_TAXID: 32046;
SOURCE 5 ORGANISM_SCIENTIFIC: SYNECHOCOCCUS ELONGATUS;
KEYWDS MEMBRANE PROTEIN, MULTIPROTEIN-PIGMENT COMPLEX,
KEYWDS 2 PHOTOSYNTHESIS
SCOP: b.34 SH3-like barrel
SCOP: b.34.4 Electron transport accessory proteins
SCOP: b.34.4.2 Photosystem I accessory protein E (PsaE)
SCOP: d.187 Photosystem I subunit PsaD
SCOP: d.187.1 Photosystem I subunit PsaD
SCOP: d.187.1.1 Photosystem I subunit PsaD
SCOP: d.58 Ferredoxin-like
SCOP: d.58.1 4Fe-4S ferredoxins
SCOP: d.58.1.2 7-Fe ferredoxin
SCOP: f.23 Single transmembrane helix
SCOP: f.23.16 Subunit III of photosystem I reaction centre, PsaF
SCOP: f.23.16.1 Subunit III of photosystem I reaction centre, PsaF
SCOP: f.23 Single transmembrane helix
SCOP: f.23.17 Subunit VIII of photosystem I reaction centre, PsaI
SCOP: f.23.17.1 Subunit VIII of photosystem I reaction centre, PsaI
SCOP: f.23 Single transmembrane helix
SCOP: f.23.18 Subunit IX of photosystem I reaction centre, PsaJ
SCOP: f.23.18.1 Subunit IX of photosystem I reaction centre, PsaJ
SCOP: f.23 Single transmembrane helix
SCOP: f.23.19 Subunit XII of photosystem I reaction centre, PsaM
SCOP: f.23.19.1 Subunit XII of photosystem I reaction centre, PsaM
SCOP: f.23 Single transmembrane helix
SCOP: f.23.20 Subunit PsaX of photosystem I reaction centre
SCOP: f.23.20.1 Subunit PsaX of photosystem I reaction centre
SCOP: f.29 Photosystem I subunits PsaA/PsaB
SCOP: f.29.1 Photosystem I subunits PsaA/PsaB
SCOP: f.29.1.1 Photosystem I subunits PsaA/PsaB
SCOP: f.30 Photosystem I reaction center subunit X, PsaK
SCOP: f.30.1 Photosystem I reaction center subunit X, PsaK
SCOP: f.30.1.1 Photosystem I reaction center subunit X, PsaK
SCOP: f.31 Photosystem I reaction center subunit XI, PsaL
SCOP: f.31.1 Photosystem I reaction center subunit XI, PsaL
SCOP: f.31.1.1 Photosystem I reaction center subunit XI, PsaL
PDB: 1q16 PDBsum
HEADER OXIDOREDUCTASE 18-JUL-03 1Q16
TITLE CRYSTAL STRUCTURE OF NITRATE REDUCTASE A, NARGHI, FROM
TITLE 2 ESCHERICHIA COLI
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
SOURCE 10 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
COMPND 4 EC: 1.7.99.4;
COMPND 9 EC: 1.7.99.4;
COMPND 15 EC: 1.7.99.4;
KEYWDS MEMBRANE PROTEIN, ELECTRON-TRANSFER, OXIDOREDUCTASE
SCOP: b.52 Double psi beta-barrel
SCOP: b.52.2 ADC-like
SCOP: b.52.2.2 Formate dehydrogenase/DMSO reductase, C-terminal domain
SCOP: c.81 Formate dehydrogenase/DMSO reductase, domains 1-3
SCOP: c.81.1 Formate dehydrogenase/DMSO reductase, domains 1-3
SCOP: c.81.1.1 Formate dehydrogenase/DMSO reductase, domains 1-3
SCOP: d.58 Ferredoxin-like
SCOP: d.58.1 4Fe-4S ferredoxins
SCOP: d.58.1.5 Ferredoxin domains from multidomain proteins
SCOP: f.21 Heme-binding four-helical bundle
SCOP: f.21.3 Respiratory nitrate reductase 1 gamma chain
SCOP: f.21.3.1 Respiratory nitrate reductase 1 gamma chain