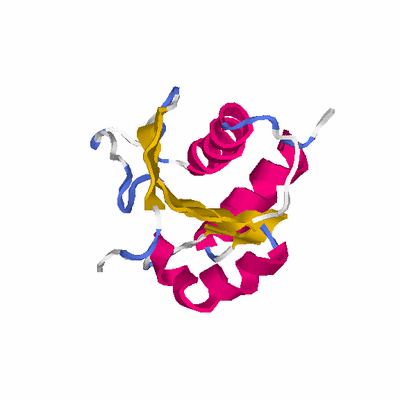
##########
# PROTEIN: Q2246_545_139.5wLII_11111_19 164 139 # Category_C1
# Length: 164
# Weight: 17652.83
# Similarity: UniProt_B6AKH2, BLAST_B6AKH2, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 68.90 M_08 1ehy_A 294 4 1e-27 15.000 113:117 (20-134:6-120)
# Evl 97.56 M_00 3ezy_A 344 4 2e-43 12.000 160:170 (1-160:91-260)
# Sid 80.49 M_23 2vf2_A 311 2 7e-24 17.000 132:146 (20-157:30-169)
# Cov 98.78 M_06 3e18_A 359 2 7e-28 11.000 162:171 (1-163:92-261)
# LaL 59.76 M_32 2h63_D 292 4 2e-06 12.000 98:100 (1-100:90-187)
# OvN 97.56 M_00 3ezy_A 344 4 2e-43 12.000 160:170 (1-160:91-260)
# OvC 77.44 M_25 1a8q_A 274 1 2e-23 13.000 127:143 (25-164:1-130)
PDB: 1ehy PDBsum
HEADER HYDROLASE 17-OCT-98 1EHY
TITLE X-RAY STRUCTURE OF THE EPOXIDE HYDROLASE FROM AGROBACTERIUM
TITLE 2 RADIOBACTER AD1
SOURCE 2 ORGANISM_SCIENTIFIC: AGROBACTERIUM TUMEFACIENS;
SOURCE 3 ORGANISM_TAXID: 358;
COMPND 4 EC: 3.3.2.3;
KEYWDS HYDROLASE, ALPHA/BETA HYDROLASE FOLD, EPOXIDE DEGRADATION,
KEYWDS 2 EPICHLOROHYDRIN
SCOP: c.69 alpha/beta-Hydrolases
SCOP: c.69.1 alpha/beta-Hydrolases
SCOP: c.69.1.11 Epoxide hydrolase
PDB: 3ezy PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 24-OCT-08 3EZY
TITLE CRYSTAL STRUCTURE OF PROBABLE DEHYDROGENASE TM_0414 FROM
TITLE 2 THERMOTOGA MARITIMA
SOURCE 2 ORGANISM_SCIENTIFIC: THERMOTOGA MARITIMA;
SOURCE 3 ORGANISM_TAXID: 2336;
KEYWDS STRUCTURAL GENOMICS, UNKNOWN FUNCTION, PSI-2, PROTEIN
KEYWDS 2 STRUCTURE INITIATIVE, NEW YORK SGX RESEARCH CENTER FOR
KEYWDS 3 STRUCTURAL GENOMICS, NYSGXRC
SCOP: none
PDB: 2vf2 PDBsum
HEADER HYDROLASE 29-OCT-07 2VF2
TITLE X-RAY CRYSTAL STRUCTURE OF HSAD FROM MYCOBACTERIUM
TITLE 2 TUBERCULOSIS
SOURCE 2 ORGANISM_SCIENTIFIC: MYCOBACTERIUM TUBERCULOSIS;
SOURCE 3 ORGANISM_TAXID: 83332;
COMPND 7 EC: 3.7.1.8;
KEYWDS META-CLEAVAGE PRODUCT HYDROLASE, HSAD, HYDROLASE, SERINE
KEYWDS 2 HYDROLASE
SCOP: none
PDB: 3e18 PDBsum
HEADER OXIDOREDUCTASE 02-AUG-08 3E18
TITLE CRYSTAL STRUCTURE OF NAD-BINDING PROTEIN FROM LISTERIA
TITLE 2 INNOCUA
SOURCE 2 ORGANISM_SCIENTIFIC: LISTERIA INNOCUA;
SOURCE 3 ORGANISM_TAXID: 1642;
KEYWDS OXIDOREDUCTASE, DEHYDROGENASE, NAD-BINDING, STRUCTURAL
KEYWDS 2 GENOMICS, PROTEIN STRUCTURE INITIATIVE, PSI, NEW YORK
KEYWDS 3 STRUCTURAL GENOMIX RESEARCH CONSORTIUM, NYSGXRC, UNKNOWN
KEYWDS 4 FUNCTION, NEW YORK SGX RESEARCH CENTER FOR STRUCTURAL
KEYWDS 5 GENOMICS
SCOP: none
PDB: 2h63 PDBsum
HEADER OXIDOREDUCTASE 30-MAY-06 2H63
TITLE CRYSTAL STRUCTURE OF HUMAN BILIVERDIN REDUCTASE A (CASP
TITLE 2 TARGET)
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 6 EC: 1.3.1.24;
KEYWDS OXIDOREDUCTASE, BILIVERDIN REDUCTASE, BLVRA, STRUCTURAL
KEYWDS 2 GENOMICS, STRUCTURAL GENOMICS CONSORTIUM, SGC
SCOP: none
PDB: 1a8q PDBsum
HEADER HALOPEROXIDASE 27-MAR-98 1A8Q
TITLE BROMOPEROXIDASE A1
SOURCE 2 ORGANISM_SCIENTIFIC: STREPTOMYCES AUREOFACIENS;
SOURCE 3 ORGANISM_TAXID: 1894;
COMPND 5 EC: 1.11.1.10;
KEYWDS HALOPEROXIDASE, OXIDOREDUCTASE
SCOP: c.69 alpha/beta-Hydrolases
SCOP: c.69.1 alpha/beta-Hydrolases
SCOP: c.69.1.12 Haloperoxidase