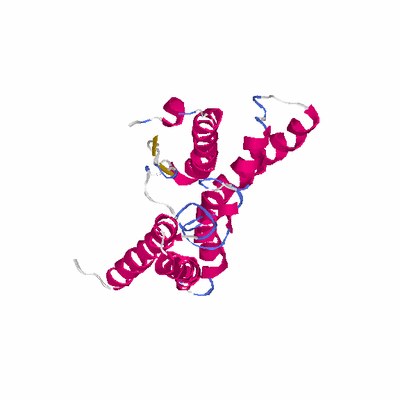
##########
# PROTEIN: Q2246_545_146.5wLII_11111_54 366 146 # Category_C1
# Length: 366
# Weight: 41530.40
# Similarity: UniProt_B6AKD7, BLAST_B6AKD7, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 51.91 M_00 3b4r_A 224 2 2e-39 20.000 190:210 (8-216:30-224)
# Evl 48.91 M_01 1jr1_A 514 2 4e-60 11.000 179:284 (88-357:1-255)
# Sid 44.54 M_00 3b4r_A 224 2 6e-05 29.000 163:190 (8-187:30-202)
# Cov 61.20 M_02 1zfj_A 491 1 2e-56 9.000 224:269 (110-364:6-243)
# LaL 33.06 M_09 1xkf_B 133 2 8e-22 16.000 121:123 (212-333:8-129)
# OvN 51.64 M_00 3b4r_A 224 2 6e-44 21.000 189:211 (8-216:30-224)
# OvC 32.51 M_39 1vr9_A 213 2 5e-09 11.000 119:160 (213-366:14-166)
PDB: 3b4r PDBsum
HEADER HYDROLASE 24-OCT-07 3B4R
TITLE SITE-2 PROTEASE FROM METHANOCALDOCOCCUS JANNASCHII
SOURCE 2 ORGANISM_SCIENTIFIC: METHANOCALDOCOCCUS JANNASCHII;
SOURCE 3 ORGANISM_TAXID: 2190;
COMPND 5 EC: 3.4.24.-;
KEYWDS INTRAMEMBRANE PROTEASE, METALLOPROTEASE, CBS DOMAIN,
KEYWDS 2 HYDROLASE, METAL-BINDING, TRANSMEMBRANE, ZINC
SCOP: none
PDB: 1jr1 PDBsum
HEADER OXIDOREDUCTASE 09-AUG-01 1JR1
TITLE CRYSTAL STRUCTURE OF INOSINE MONOPHOSPHATE DEHYDROGENASE IN
TITLE 2 COMPLEX WITH MYCOPHENOLIC ACID
SOURCE 2 ORGANISM_SCIENTIFIC: CRICETULUS GRISEUS;
SOURCE 3 ORGANISM_COMMON: CHINESE HAMSTER;
SOURCE 4 ORGANISM_TAXID: 10029;
COMPND 5 EC: 1.1.1.205;
KEYWDS DEHYDROGENASE, IMPD, IMPDH, GUANINE NUCLEOTIDE SYNTHESIS,
KEYWDS 2 MYCOPHENOLIC ACID, MPA, OXIDOREDUCTASE
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.5 Inosine monophosphate dehydrogenase (IMPDH)
SCOP: c.1.5.1 Inosine monophosphate dehydrogenase (IMPDH)
SCOP: d.37 CBS-domain
SCOP: d.37.1 CBS-domain
SCOP: d.37.1.1 CBS-domain
PDB: 1zfj PDBsum
HEADER OXIDOREDUCTASE 29-MAR-99 1ZFJ
TITLE INOSINE MONOPHOSPHATE DEHYDROGENASE (IMPDH; EC 1.1.1.205)
TITLE 2 FROM STREPTOCOCCUS PYOGENES
SOURCE 2 ORGANISM_SCIENTIFIC: STREPTOCOCCUS PYOGENES;
SOURCE 3 ORGANISM_TAXID: 1314;
COMPND 5 EC: 1.1.1.205;
KEYWDS IMPDH, DEHYDROGENASE, CBS DOMAINS, OXIDOREDUCTASE
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.5 Inosine monophosphate dehydrogenase (IMPDH)
SCOP: c.1.5.1 Inosine monophosphate dehydrogenase (IMPDH)
SCOP: d.37 CBS-domain
SCOP: d.37.1 CBS-domain
SCOP: d.37.1.1 CBS-domain
PDB: 1xkf PDBsum
HEADER UNKNOWN FUNCTION 28-SEP-04 1XKF
TITLE CRYSTAL STRUCTURE OF HYPOXIC RESPONSE PROTEIN I (HRPI) WITH
TITLE 2 TWO COORDINATED ZINC IONS
SOURCE 2 ORGANISM_SCIENTIFIC: MYCOBACTERIUM TUBERCULOSIS;
SOURCE 3 ORGANISM_TAXID: 1773;
KEYWDS CBS DOMAIN, UNKNOWN FUNCTION
SCOP: d.37 CBS-domain
SCOP: d.37.1 CBS-domain
SCOP: d.37.1.1 CBS-domain
PDB: 1vr9 PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 17-FEB-05 1VR9
TITLE CRYSTAL STRUCTURE OF CBS DOMAIN PROTEIN/ACT DOMAIN PROTEIN
TITLE 2 (TM0892) FROM THERMOTOGA MARITIMA AT 1.70 A RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: THERMOTOGA MARITIMA;
SOURCE 3 ORGANISM_TAXID: 2336;
KEYWDS TM0892, CBS DOMAIN PROTEIN/ACT DOMAIN PROTEIN, STRUCTURAL
KEYWDS 2 GENOMICS, JOINT CENTER FOR STRUCTURAL GENOMICS, JCSG,
KEYWDS 3 PROTEIN STRUCTURE INITIATIVE, PSI, UNKNOWN FUNCTION
SCOP: d.37 CBS-domain
SCOP: d.37.1 CBS-domain
SCOP: d.37.1.1 CBS-domain