##########
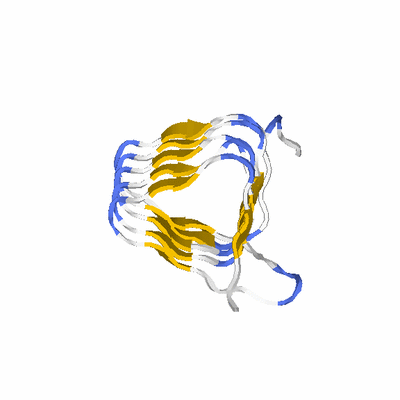
# PROTEIN: Q2246_545_160.5wLII_11172_11 132 160 # Category_C1
# Length: 132
# Weight: 14420.40
# Similarity: UniProt_B6AN64, BLAST_B6AN64, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 68.18 M_00 1hv9_B 456 8 2e-08 17.000 90:98 (19-113:258-350)
# Evl 93.94 M_00 1hv9_B 456 8 2e-31 10.000 124:138 (5-128:244-381)
# Sid 32.58 M_04 2i5k_B 488 2 0.056 25.000 43:47 (71-113:439-485)
# Cov 94.70 M_01 z0kx 458 0 1e-29 9.000 125:139 (4-128:242-380)
# LaL 38.64 M_03 2f9c_A 334 2 0.022 15.000 51:51 (71-121:73-123)
# OvN 90.15 M_12 z11z 193 0 5e-06 10.000 119:133 (1-128:18-146)
# OvC 68.18 M_09 z0kx 458 0 0.054 12.000 90:94 (39-132:261-350)
PDB: 1hv9 PDBsum
HEADER TRANSFERASE 08-JAN-01 1HV9
TITLE STRUCTURE OF E. COLI GLMU: ANALYSIS OF PYROPHOSPHORYLASE
TITLE 2 AND ACETYLTRANSFERASE ACTIVE SITES
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 5 EC: 2.7.7.23;
KEYWDS LEFT-HANDED PARALLEL BETA-HELIX, TRANSFERASE
SCOP: b.81 Single-stranded left-handed beta-helix
SCOP: b.81.1 Trimeric LpxA-like enzymes
SCOP: b.81.1.4 GlmU C-terminal domain-like
SCOP: c.68 Nucleotide-diphospho-sugar transferases
SCOP: c.68.1 Nucleotide-diphospho-sugar transferases
SCOP: c.68.1.5 UDP-glucose pyrophosphorylase
PDB: 2i5k PDBsum
HEADER TRANSFERASE 25-AUG-06 2I5K
TITLE CRYSTAL STRUCTURE OF UGP1P
SOURCE 2 ORGANISM_SCIENTIFIC: SACCHAROMYCES CEREVISIAE;
SOURCE 3 ORGANISM_COMMON: BAKER'S YEAST;
SOURCE 4 ORGANISM_TAXID: 4932;
COMPND 7 EC: 2.7.7.9
KEYWDS LEFT-HANDED BETA-HELIX, SGC DOMAIN, TRANSFERASE
SCOP: none
PDB: 2f9c PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 05-DEC-05 2F9C
TITLE CRYSTAL STRUCTURE OF YDCK FROM SALMONELLA CHOLERAE. NESG
TITLE 2 TARGET SCR6
SOURCE 2 ORGANISM_SCIENTIFIC: SALMONELLA ENTERICA;
SOURCE 3 ORGANISM_TAXID: 28901;
KEYWDS STRUCTURAL GENOMICS, UNKNOWN FUNCTION, PSI, PROTEIN
KEYWDS 2 STRUCTURE INITIATIVE, NORTHEAST STRUCTURAL GENOMICS
KEYWDS 3 CONSORTIUM, NESG
SCOP: b.81 Single-stranded left-handed beta-helix
SCOP: b.81.1 Trimeric LpxA-like enzymes
SCOP: b.81.1.7 YdcK-like
PDB: 2eg0 PDBsum
HEADER STRUCTURAL GENOMICS UNKNOWN FUNCTION 27-FEB-07 2EG0
TITLE CRYSTAL STRUCTURE OF HYPOTHETICAL PROTEIN(GK2848) FROM
TITLE 2 GEOBACILLUS KAUSTOPHILUS
SOURCE 2 ORGANISM_SCIENTIFIC: GEOBACILLUS KAUSTOPHILUS;
SOURCE 3 ORGANISM_TAXID: 1462;
KEYWDS HYPOTHETICAL PROTEIN, GEOBACILLUS KAUSTOPHILUS, STRUCTURAL
KEYWDS 2 GENOMICS, NPPSFA, NATIONAL PROJECT ON PROTEIN STRUCTURAL
KEYWDS 3 AND FUNCTIONAL ANALYSES, RIKEN STRUCTURAL
KEYWDS 4 GENOMICS/PROTEOMICS INITIATIVE, RSGI, STRUCTURAL GENOMICS
KEYWDS 5 UNKNOWN FUNCTION
SCOP: none