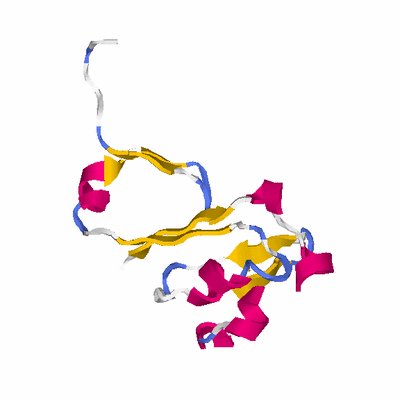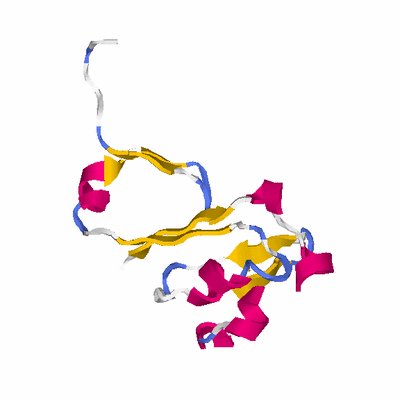##########
# PROTEIN: Q2246_545_169.5wLII_11172_58 101 169 # Category_B
# Length: 101
# Weight: 11166.97
# Similarity: UniProt_B6ANA8, BLAST_B6ANA8, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 87.13 M_00 3bk7_A 607 1 4e-38 22.000 88:98 (3-94:20-113)
# Evl 87.13 M_00 3bk7_A 607 1 4e-38 22.000 88:98 (3-94:20-113)
# Sid 52.48 M_07 1bqx_A 77 2 0.014 32.000 53:56 (5-60:1-53)
# Cov 95.05 M_01 1h7x_B 1025 4 7e-11 14.000 96:103 (5-100:72-174)
# LaL 53.47 M_24 1jnr_B 150 12 4e-05 24.000 54:54 (10-63:11-64)
# OvN 86.14 M_00 1y4z_B 512 1 1e-28 14.000 87:92 (1-92:177-263)
# OvC 94.06 M_00 3bk7_A 607 1 5e-22 18.000 95:111 (3-101:20-126)
PDB: 3bk7 PDBsum
HEADER HYDROLYASE/TRANSLATION 06-DEC-07 3BK7
TITLE STRUCTURE OF THE COMPLETE ABCE1/RNAASE-L INHIBITOR PROTEIN
TITLE 2 FROM PYROCOCCUS ABYSII
SOURCE 2 ORGANISM_SCIENTIFIC: PYROCOCCUS ABYSSI;
SOURCE 3 ORGANISM_TAXID: 29292;
KEYWDS ABC ATPASE, IRON-SULFUR CLUSTER, ADENOSINE DIPHOSPHATE, ATP-
KEYWDS 2 BINDING, NUCLEOTIDE-BINDING, HYDROLYASE/TRANSLATION COMPLEX
SCOP: none
PDB: 1bqx PDBsum
HEADER ELECTRON TRANSPORT 20-AUG-98 1BQX
TITLE ARTIFICIAL FE8S8 FERREDOXIN: THE D13C VARIANT OF BACILLUS
TITLE 2 SCHLEGELII FE7S8 FERREDOXIN
SOURCE 2 ORGANISM_SCIENTIFIC: BACILLUS SCHLEGELII;
SOURCE 3 ORGANISM_TAXID: 1484;
KEYWDS IRON-SULFUR PROTEIN, ELECTRON TRANSPORT
SCOP: d.58 Ferredoxin-like
SCOP: d.58.1 4Fe-4S ferredoxins
SCOP: d.58.1.2 7-Fe ferredoxin
PDB: 1h7x PDBsum
HEADER ELECTRON TRANSFER 19-JAN-01 1H7X
TITLE DIHYDROPYRIMIDINE DEHYDROGENASE (DPD) FROM PIG, TERNARY
TITLE 2 COMPLEX OF A MUTANT ENZYME (C671A), NADPH AND
TITLE 3 5-FLUOROURACIL
SOURCE 2 ORGANISM_SCIENTIFIC: SUS SCROFA;
SOURCE 3 ORGANISM_COMMON: WILD BOAR;
SOURCE 4 ORGANISM_TAXID: 9823;
COMPND 6 EC: 1.3.1.2;
KEYWDS ELECTRON TRANSFER, FLAVIN, IRON-SULFUR CLUSTERS, PYRIMIDINE
KEYWDS 2 CATABOLISM, 5-FLUOROURACIL DEGRADATION, OXIDOREDUCTASE
SCOP: a.1 Globin-like
SCOP: a.1.2 alpha-helical ferredoxin
SCOP: a.1.2.2 Dihydropyrimidine dehydrogenase, N-terminal domain
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.4 FMN-linked oxidoreductases
SCOP: c.1.4.1 FMN-linked oxidoreductases
SCOP: c.3 FAD/NAD(P)-binding domain
SCOP: c.3.1 FAD/NAD(P)-binding domain
SCOP: c.3.1.1 C-terminal domain of adrenodoxin reductase-like
SCOP: c.4 Nucleotide-binding domain
SCOP: c.4.1 Nucleotide-binding domain
SCOP: c.4.1.1 N-terminal domain of adrenodoxin reductase-like
SCOP: d.58 Ferredoxin-like
SCOP: d.58.1 4Fe-4S ferredoxins
SCOP: d.58.1.5 Ferredoxin domains from multidomain proteins
PDB: 1jnr PDBsum
HEADER OXIDOREDUCTASE 25-JUL-01 1JNR
TITLE STRUCTURE OF ADENYLYLSULFATE REDUCTASE FROM THE
TITLE 2 HYPERTHERMOPHILIC ARCHAEOGLOBUS FULGIDUS AT 1.6 RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: ARCHAEOGLOBUS FULGIDUS DSM 4304;
SOURCE 3 ORGANISM_TAXID: 224325;
SOURCE 6 ORGANISM_SCIENTIFIC: ARCHAEOGLOBUS FULGIDUS DSM 4304;
COMPND 5 EC: 1.8.99.2;
COMPND 10 EC: 1.8.99.2
KEYWDS SULFUR METABOLISM/ ADENYLYLSULFATE REDUCTASE/ IRON-SULFUR
KEYWDS 2 FLAVOPROTEIN/ CRYSTAL STRUCTURE/CATALYSIS, OXIDOREDUCTASE
SCOP: a.7 Spectrin repeat-like
SCOP: a.7.3 Succinate dehydrogenase/fumarate reductase flavoprotein C-terminal domain
SCOP: a.7.3.1 Succinate dehydrogenase/fumarate reductase flavoprotein C-terminal domain
SCOP: c.3 FAD/NAD(P)-binding domain
SCOP: c.3.1 FAD/NAD(P)-binding domain
SCOP: c.3.1.4 Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain
SCOP: d.58 Ferredoxin-like
SCOP: d.58.1 4Fe-4S ferredoxins
SCOP: d.58.1.5 Ferredoxin domains from multidomain proteins
SCOP: d.168 Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
SCOP: d.168.1 Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
SCOP: d.168.1.1 Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
PDB: 1y4z PDBsum
HEADER OXIDOREDUCTASE 01-DEC-04 1Y4Z
TITLE THE CRYSTAL STRUCTURE OF NITRATE REDUCTASE A, NARGHI, IN
TITLE 2 COMPLEX WITH THE Q-SITE INHIBITOR PENTACHLOROPHENOL
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
SOURCE 11 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
COMPND 6 EC: 1.7.99.4;
COMPND 13 EC: 1.7.99.4;
COMPND 21 EC: 1.7.99.4;
KEYWDS NITRATE REDUCTION, ELECTRON TRANSFER, MEMBRANE PROTEIN, Q-
KEYWDS 2 SITE, OXIDOREDUCTASE
SCOP: b.52 Double psi beta-barrel
SCOP: b.52.2 ADC-like
SCOP: b.52.2.2 Formate dehydrogenase/DMSO reductase, C-terminal domain
SCOP: c.81 Formate dehydrogenase/DMSO reductase, domains 1-3
SCOP: c.81.1 Formate dehydrogenase/DMSO reductase, domains 1-3
SCOP: c.81.1.1 Formate dehydrogenase/DMSO reductase, domains 1-3
SCOP: f.21 Heme-binding four-helical bundle
SCOP: f.21.3 Respiratory nitrate reductase 1 gamma chain
SCOP: f.21.3.1 Respiratory nitrate reductase 1 gamma chain