##########
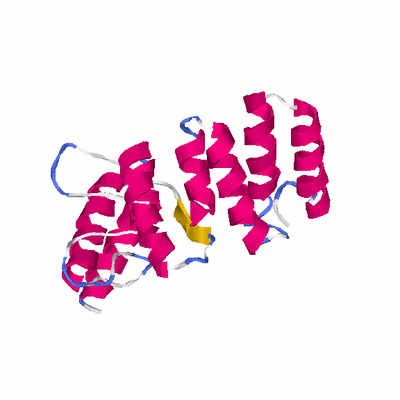
# PROTEIN: Q2246_545_183.5wLII_11181_54 216 183 # Category_C1
# Length: 216
# Weight: 25621.65
# Similarity: UniProt_B6AMT0, BLAST_B6AMT0, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 62.96 M_01 2fo7_A 136 2 4e-28 17.000 136:167 (26-192:1-136)
# Evl 94.91 M_00 1w3b_A 388 2 2e-40 13.000 205:222 (5-215:148-363)
# Sid 29.17 M_39 1hh8_A 213 1 1e-06 19.000 63:66 (121-186:36-98)
# Cov 96.76 M_10 2gw1_A 514 2 6e-19 12.000 209:230 (3-215:248-473)
# LaL 29.63 M_38 z0sf 110 0 1e-06 15.000 64:64 (7-70:36-99)
# OvN 93.52 M_00 1w3b_A 388 2 5e-33 10.000 202:220 (1-210:177-388)
# OvC 74.07 M_02 3edt_B 283 4 5e-23 12.000 160:183 (45-216:20-190)
PDB: 2fo7 PDBsum
HEADER DE NOVO PROTEIN 12-JAN-06 2FO7
TITLE CRYSTAL STRUCTURE OF AN 8 REPEAT CONSENSUS TPR SUPERHELIX
TITLE 2 (TRIGONAL CRYSTAL FORM)
KEYWDS TETRATRICOPEPTIDE REPEAT, TPR, CONSENSUS PROTEIN,
KEYWDS 2 SUPERHELIX, DE NOVO PROTEIN
SCOP: k.38 TPR domain-based design
SCOP: k.38.1 TPR domain-based design
SCOP: k.38.1.1 TPR domain-based design
PDB: 1w3b PDBsum
HEADER TRANSFERASE 14-JUL-04 1W3B
TITLE THE SUPERHELICAL TPR DOMAIN OF O-LINKED GLCNAC TRANSFERASE
TITLE 2 REVEALS STRUCTURAL SIMILARITIES TO IMPORTIN ALPHA.
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 7 EC: 2.4.1.-;
KEYWDS OGT, GLCNAC, NUCLEOPORIN, O-LINKED GLYCOSYLATION, TPR
KEYWDS 2 REPEAT, PROTEIN BINDING, SIGNAL TRANSDUCTION, TRANSFERASE
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)
PDB: 1hh8 PDBsum
HEADER CELL CYCLE 21-DEC-00 1HH8
TITLE THE ACTIVE N-TERMINAL REGION OF P67PHOX: STRUCTURE AT
TITLE 2 1.8 ANGSTROM RESOLUTION AND BIOCHEMICAL CHARACTERIZATIONS
TITLE 3 OF THE A128V MUTANT IMPLICATED IN CHRONIC GRANULOMATOUS
TITLE 4 DISEASE
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
KEYWDS CELL CYCLE, PHAGOCYTE OXIDASE FACTOR, SH3 DOMAIN, REPEAT,
KEYWDS 2 TPR REPEAT CELL CYCLE
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)
PDB: 2gw1 PDBsum
HEADER PROTEIN TRANSPORT 03-MAY-06 2GW1
TITLE CRYSTAL STRUCTURE OF THE YEAST TOM70
SOURCE 2 ORGANISM_SCIENTIFIC: SACCHAROMYCES CEREVISIAE;
SOURCE 3 ORGANISM_COMMON: BAKER'S YEAST;
SOURCE 4 ORGANISM_TAXID: 4932;
KEYWDS TPR, PROTEIN TRANSPORT
SCOP: none
PDB: 3edt PDBsum
HEADER MOTOR PROTEIN, TRANSPORT PROTEIN 03-SEP-08 3EDT
TITLE CRYSTAL STRUCTURE OF THE MUTATED S328N HKLC2 TPR DOMAIN
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
KEYWDS SUPERHELICAL, STRUCTURAL GENOMICS, STRUCTURAL GENOMICS
KEYWDS 2 CONSORTIUM, SGC, COILED COIL, MICROTUBULE, MOTOR PROTEIN,
KEYWDS 3 PHOSPHOPROTEIN, POLYMORPHISM, TPR REPEAT, TRANSPORT PROTEIN
SCOP: none