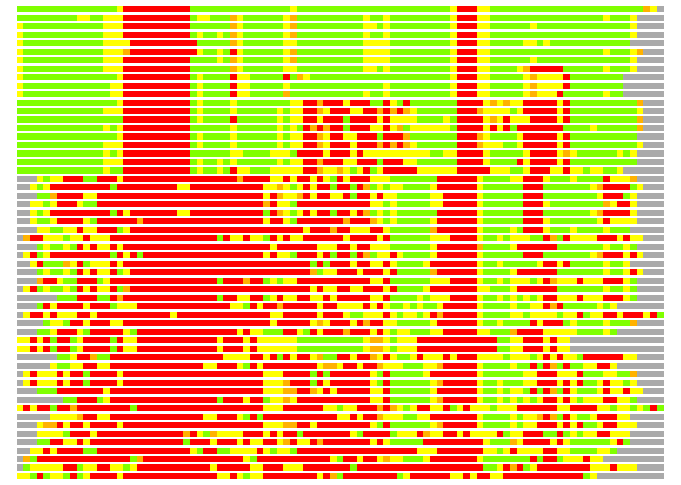
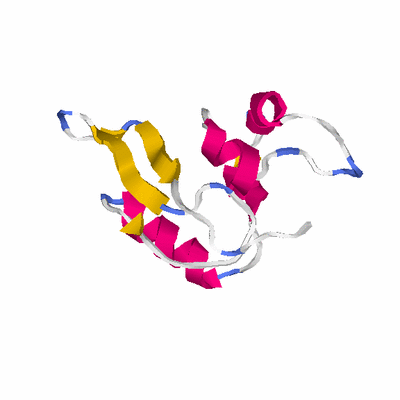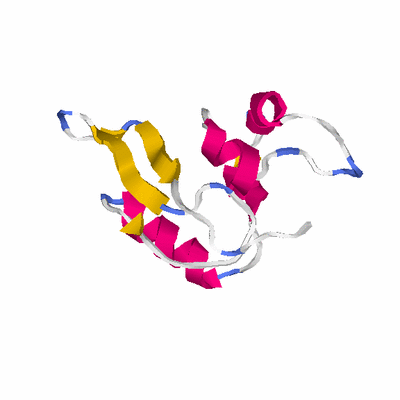##########
# PROTEIN: Q2246_545_188.5wLII_11181_70 137 188 # Category_C1
# Length: 137
# Weight: 15127.11
# Similarity: UniProt_B6AMU6, BLAST_B6AMU6, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 61.31 M_01 2gj3_A 120 2 3e-14 17.000 84:104 (1-94:25-118)
# Evl 76.64 M_00 3eqv_A 542 2 3e-21 11.000 105:144 (2-134:226-341)
# Sid 34.31 M_00 3c4s_A 66 3 0.010 38.000 47:50 (68-117:4-50)
# Cov 85.40 M_12 2jch_A 720 5 3e-05 16.000 117:133 (3-131:354-474)
# LaL 22.63 M_25 1e3u_D 246 1 0.040 16.000 31:31 (66-96:43-73)
# OvN 61.31 M_01 2gj3_A 120 2 3e-14 17.000 84:104 (1-94:25-118)
# OvC 76.64 M_00 3eqv_A 542 2 3e-21 11.000 105:144 (2-134:226-341)
PDB: 2gj3 PDBsum
HEADER TRANSFERASE 30-MAR-06 2GJ3
TITLE CRYSTAL STRUCTURE OF THE FAD-CONTAINING PAS DOMAIN OF THE
TITLE 2 PROTEIN NIFL FROM AZOTOBACTER VINELANDII.
SOURCE 2 ORGANISM_SCIENTIFIC: AZOTOBACTER VINELANDII;
SOURCE 3 ORGANISM_TAXID: 354;
COMPND 5 EC: 2.7.13.3;
KEYWDS PAS DOMAIN, FAD, REDOX SENSOR, ATOMIC RESOLUTION,
KEYWDS 2 TRANSFERASE
SCOP: none
PDB: 3eqv PDBsum
HEADER BIOSYNTHETIC PROTEIN 01-OCT-08 3EQV
TITLE CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 2 FROM
TITLE 2 NEISSERIA GONORRHOEAE CONTAINING FOUR MUTATIONS ASSOCIATED
TITLE 3 WITH PENICILLIN RESISTANCE
SOURCE 2 ORGANISM_SCIENTIFIC: NEISSERIA GONORRHOEAE;
SOURCE 3 ORGANISM_TAXID: 485;
COMPND 5 EC: 3.4.16.4;
KEYWDS PENICILLIN-BINDING PROTEIN, CLASS B TRANSPEPTIDASE,
KEYWDS 2 PENICILLIN RESISTANCE, CELL DIVISION, CELL INNER MEMBRANE,
KEYWDS 3 CELL MEMBRANE, CELL SHAPE, CELL WALL BIOGENESIS/DEGRADATION,
KEYWDS 4 PEPTIDOGLYCAN SYNTHESIS, BIOSYNTHETIC PROTEIN
SCOP: none
PDB: 3c4s PDBsum
HEADER STRUCTURAL GENOMICS, UNKNOWN FUNCTION 30-JAN-08 3C4S
TITLE CRYSTAL STRUCTURE OF THE SSL0352 PROTEIN FROM SYNECHOCYSTIS
TITLE 2 SP. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET SGR42
SOURCE 2 ORGANISM_SCIENTIFIC: SYNECHOCYSTIS SP. PCC 6803;
SOURCE 3 ORGANISM_TAXID: 1148;
KEYWDS P74795_SYNY3, SSL0352, NESG, SGR42, STRUCTURAL GENOMICS,
KEYWDS 2 PSI-2, PROTEIN STRUCTURE INITIATIVE, NORTHEAST STRUCTURAL
KEYWDS 3 GENOMICS CONSORTIUM, UNKNOWN FUNCTION
SCOP: none
PDB: 2jch PDBsum
HEADER DRUG-BINDING PROTEIN 23-DEC-06 2JCH
TITLE STRUCTURAL AND MECHANISTIC BASIS OF PENICILLIN BINDING
TITLE 2 PROTEIN INHIBITION BY LACTIVICINS
SOURCE 2 ORGANISM_SCIENTIFIC: STREPTOCOCCUS PNEUMONIAE;
SOURCE 3 ORGANISM_TAXID: 171101;
KEYWDS PEPTIDOGLYCAN SYNTHESIS MULTIFUNCTIONAL ENZYME, CELL WALL,
KEYWDS 2 PEPTIDOGLYCAN, GAMMA LACTAM ANTIBIOTICS, BINDING PROTEIN,
KEYWDS 3 DRUG-BINDING PROTEIN
SCOP: none
PDB: 1e3u PDBsum
HEADER BETA-LACTAMASE 23-JUN-00 1E3U
TITLE MAD STRUCTURE OF OXA10 CLASS D BETA-LACTAMASE
SOURCE 2 ORGANISM_SCIENTIFIC: PSEUDOMONAS AERUGINOSA;
SOURCE 3 ORGANISM_TAXID: 287;
COMPND 4 EC: 3.5.2.6;
KEYWDS BETA-LACTAMASE, ANTIOBITIC RESISTANCE
SCOP: e.3 beta-lactamase/transpeptidase-like
SCOP: e.3.1 beta-lactamase/transpeptidase-like
SCOP: e.3.1.1 beta-Lactamase/D-ala carboxypeptidase