##########
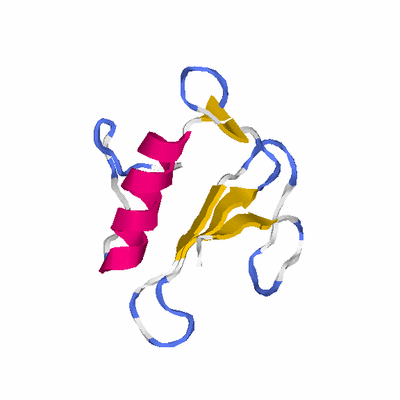
# PROTEIN: Q2246_545_19.5wLII_10580_1 106 19 # Category_C1
# Length: 106
# Weight: 11527.43
# Similarity: UniProt_A3EUW0, BLAST_A3EUW0, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 72.64 M_00 1jk3_A 158 1 0.016 18.000 77:77 (29-105:57-133)
# Evl 59.43 M_00 1y9q_A 192 1 2e-15 11.000 63:63 (39-101:11-73)
# Sid 19.81 M_34 2ras_B 212 2 0.056 23.000 21:21 (51-71:30-50)
# Cov 72.64 M_00 1jk3_A 158 1 0.016 18.000 77:77 (29-105:57-133)
# LaL 72.64 M_00 1jk3_A 158 1 0.016 18.000 77:77 (29-105:57-133)
# OvN 51.89 M_35 3c64_A 179 1 0.067 14.000 55:62 (3-64:78-132)
# OvC 58.49 M_20 2bnm_A 198 20 5e-06 13.000 62:68 (41-106:11-74)
PDB: 1jk3 PDBsum
HEADER HYDROLASE 11-JUL-01 1JK3
TITLE CRYSTAL STRUCTURE OF HUMAN MMP-12 (MACROPHAGE ELASTASE) AT
TITLE 2 TRUE ATOMIC RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 7 EC: 3.4.24.65;
KEYWDS MATRIX METALLOPROTEINASE, BATIMASTAT, BB94, HYDROXAMIC ACID,
KEYWDS 2 MMP12, ELASTASE, COMPLEX (ELASTASE/INHIBITOR), METALLO
KEYWDS 3 ELASTASE, HYDROLASE
SCOP: d.92 Zincin-like
SCOP: d.92.1 Metalloproteases ("zincins"), catalytic domain
SCOP: d.92.1.11 Matrix metalloproteases, catalytic domain
PDB: 1y9q PDBsum
HEADER TRANSCRIPTION REGULATOR 16-DEC-04 1Y9Q
TITLE CRYSTAL STRUCTURE OF HTH_3 FAMILY TRANSCRIPTIONAL REGULATOR
TITLE 2 FROM VIBRIO CHOLERAE
SOURCE 2 ORGANISM_SCIENTIFIC: VIBRIO CHOLERAE;
SOURCE 3 ORGANISM_TAXID: 666;
KEYWDS HTH_3 FAMILY, TRANSCRIPTIONAL REGULAATOR, STRUCUTRAL
KEYWDS 2 GENOMICS, PROTEIN STRUCTURE INITIATIVE, PSI, NEW YORK SGX
KEYWDS 3 RESEARCH CENTER FOR STRUCTURAL GENOMICS, NYSGXRC, DIMER,
KEYWDS 4 TWO-DOMAIN STRUCTURE, STRUCTURAL GENOMICS, TRANSCRIPTION
KEYWDS 5 REGULATOR
SCOP: a.35 lambda repressor-like DNA-binding domains
SCOP: a.35.1 lambda repressor-like DNA-binding domains
SCOP: a.35.1.8 Probable transcriptional regulator VC1968, N-terminal domain
SCOP: b.82 Double-stranded beta-helix
SCOP: b.82.1 RmlC-like cupins
SCOP: b.82.1.15 Probable transcriptional regulator VC1968, C-terminal domain
PDB: 2ras PDBsum
HEADER TRANSCRIPTION 17-SEP-07 2RAS
TITLE CRYSTAL STRUCTURE OF PREDICTED TRANSCRIPTIONAL REGULATOR OF
TITLE 2 TETR/ACRR FAMILY (YP_495839.1) FROM NOVOSPHINGOBIUM
TITLE 3 AROMATICIVORANS DSM 12444 AT 1.80 A RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: NOVOSPHINGOBIUM AROMATICIVORANS DSM
SOURCE 4 ORGANISM_TAXID: 279238;
KEYWDS YP_495839.1, PREDICTED TRANSCRIPTIONAL REGULATOR OF
KEYWDS 2 TETR/ACRR FAMILY, BACTERIAL REGULATORY PROTEINS, TETR
KEYWDS 3 FAMILY, STRUCTURAL GENOMICS, JOINT CENTER FOR STRUCTURAL
KEYWDS 4 GENOMICS, JCSG, PROTEIN STRUCTURE INITIATIVE, PSI-2, DNA-
KEYWDS 5 BINDING, TRANSCRIPTION REGULATION
SCOP: none
PDB: 3c64 PDBsum
HEADER CD36-BINDING PROTEIN,CELL ADHESION 02-FEB-08 3C64
TITLE THE MC179 PORTION OF THE CYSTEINE-RICH INTERDOMAIN REGION
TITLE 2 (CIDR) OF A PLASMODIUM FALCIPARUM ERYTHROCYTE MEMBRANE
TITLE 3 PROTEIN-1 (PFEMP1)
SOURCE 2 ORGANISM_SCIENTIFIC: PLASMODIUM FALCIPARUM;
KEYWDS ALPHA-HELICAL, THREE-HELIX BUNDLE, CD36-BINDING PROTEIN,
KEYWDS 2 CELL ADHESION, CD36-BINDING PROTEIN,CELL ADHESION
SCOP: none
PDB: 2bnm PDBsum
HEADER OXIDOREDUCTASE 29-MAR-05 2BNM
TITLE STRUCTURE OF A ZN ENZYME
SOURCE 2 ORGANISM_SCIENTIFIC: STREPTOMYCES WEDMORENSIS;
SOURCE 3 ORGANISM_TAXID: 43759;
KEYWDS OXIDOREDUCTASE, EPOXIDASE, CUPIN, HTH, CATION-DEPENDANT,
KEYWDS 2 ZINC, FOSFOMYCIN
SCOP: a.35 lambda repressor-like DNA-binding domains
SCOP: a.35.1 lambda repressor-like DNA-binding domains
SCOP: a.35.1.3 SinR domain-like
SCOP: b.82 Double-stranded beta-helix
SCOP: b.82.1 RmlC-like cupins
SCOP: b.82.1.10 TM1459-like