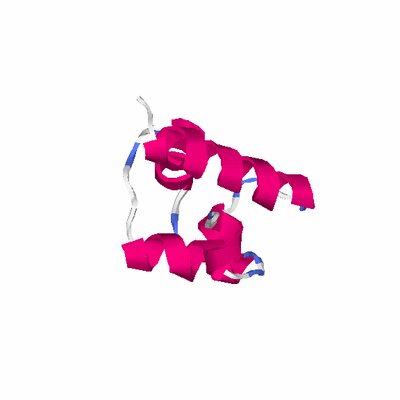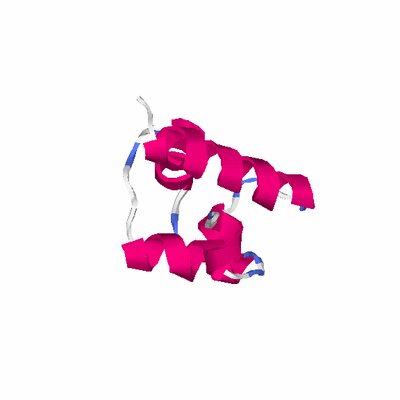##########
# PROTEIN: Q2246_545_193.5wLII_11181_81 97 193 # Category_C1
# Length: 97
# Weight: 10498.16
# Similarity: UniProt_B6AMV7, BLAST_B6AMV7, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 63.92 M_01 1y7y_A 74 2 2e-14 25.000 62:64 (32-93:10-73)
# Evl 62.89 M_00 1y9q_A 192 1 1e-15 20.000 61:63 (33-93:9-71)
# Sid 56.70 M_11 3clc_B 82 4 5e-09 26.000 55:57 (40-94:16-72)
# Cov 82.47 M_38 2jvl_A 107 1 0.002 13.000 80:94 (7-89:2-92)
# LaL 50.52 M_15 3bs3_A 76 1 7e-05 20.000 49:49 (45-93:22-70)
# OvN 77.32 M_37 2jvl_A 107 1 3e-04 13.000 75:91 (3-89:14-92)
# OvC 74.23 M_07 3f52_E 117 8 6e-09 18.000 72:88 (12-97:5-92)
PDB: 1y7y PDBsum
HEADER TRANSCRIPTION REGULATOR 10-DEC-04 1Y7Y
TITLE HIGH-RESOLUTION CRYSTAL STRUCTURE OF THE RESTRICTION-
TITLE 2 MODIFICATION CONTROLLER PROTEIN C.AHDI FROM AEROMONAS
TITLE 3 HYDROPHILA
SOURCE 2 ORGANISM_SCIENTIFIC: AEROMONAS HYDROPHILA;
SOURCE 3 ORGANISM_TAXID: 644;
KEYWDS HELIX-TURN-HELIX, DNA-BINDING PROTEIN, TRANSCRIPTIONAL
KEYWDS 2 REGULATOR, TRANSCRIPTION REGULATOR
SCOP: a.35 lambda repressor-like DNA-binding domains
SCOP: a.35.1 lambda repressor-like DNA-binding domains
SCOP: a.35.1.3 SinR domain-like
PDB: 1y9q PDBsum
HEADER TRANSCRIPTION REGULATOR 16-DEC-04 1Y9Q
TITLE CRYSTAL STRUCTURE OF HTH_3 FAMILY TRANSCRIPTIONAL REGULATOR
TITLE 2 FROM VIBRIO CHOLERAE
SOURCE 2 ORGANISM_SCIENTIFIC: VIBRIO CHOLERAE;
SOURCE 3 ORGANISM_TAXID: 666;
KEYWDS HTH_3 FAMILY, TRANSCRIPTIONAL REGULAATOR, STRUCUTRAL
KEYWDS 2 GENOMICS, PROTEIN STRUCTURE INITIATIVE, PSI, NEW YORK SGX
KEYWDS 3 RESEARCH CENTER FOR STRUCTURAL GENOMICS, NYSGXRC, DIMER,
KEYWDS 4 TWO-DOMAIN STRUCTURE, STRUCTURAL GENOMICS, TRANSCRIPTION
KEYWDS 5 REGULATOR
SCOP: a.35 lambda repressor-like DNA-binding domains
SCOP: a.35.1 lambda repressor-like DNA-binding domains
SCOP: a.35.1.8 Probable transcriptional regulator VC1968, N-terminal domain
SCOP: b.82 Double-stranded beta-helix
SCOP: b.82.1 RmlC-like cupins
SCOP: b.82.1.15 Probable transcriptional regulator VC1968, C-terminal domain
PDB: 3clc PDBsum
HEADER TRANSCRIPTION REGULATOR/DNA 18-MAR-08 3CLC
TITLE CRYSTAL STRUCTURE OF THE RESTRICTION-MODIFICATION
TITLE 2 CONTROLLER PROTEIN C.ESP1396I TETRAMER IN COMPLEX WITH ITS
TITLE 3 NATURAL 35 BASE-PAIR OPERATOR
SOURCE 2 ORGANISM_SCIENTIFIC: ENTEROBACTER SP. RFL1396;
KEYWDS PROTEIN-DNA COMPLEX, TRANSCRIPTIONAL REGULATOR, HELIX-TURN-
KEYWDS 2 HELIX, DNA-BENDING, PLASMID, TRANSCRIPTION REGULATOR/DNA
KEYWDS 3 COMPLEX
SCOP: none
PDB: 2jvl PDBsum
HEADER TRANSCRIPTION 20-SEP-07 2JVL
TITLE NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF MBF1 OF
TITLE 2 TRICHODERMA REESEI
SOURCE 2 ORGANISM_SCIENTIFIC: TRICHODERMA REESEI;
KEYWDS MBF1, COACTIVATOR, TRICHODERMA REESEI, HELIX-TURN-HELIX,
KEYWDS 2 PROTEIN BINDING, TRANSCRIPTION
SCOP: none
PDB: 3bs3 PDBsum
HEADER DNA BINDING PROTEIN 21-DEC-07 3BS3
TITLE CRYSTAL STRUCTURE OF A PUTATIVE DNA-BINDING PROTEIN FROM
TITLE 2 BACTEROIDES FRAGILIS
SOURCE 2 ORGANISM_SCIENTIFIC: BACTEROIDES FRAGILIS NCTC 9343;
SOURCE 3 ORGANISM_TAXID: 272559;
KEYWDS DNA-BINDING, XRE-FAMILY, STRUCTURAL GENOMICS, PSI-2,
KEYWDS 2 PROTEIN STRUCTURE INITIATIVE, MIDWEST CENTER FOR STRUCTURAL
KEYWDS 3 GENOMICS, MCSG, DNA BINDING PROTEIN
SCOP: none
PDB: 3f52 PDBsum
HEADER TRANSCRIPTION ACTIVATOR 03-NOV-08 3F52
TITLE CRYSTAL STRUCTURE OF THE CLP GENE REGULATOR CLGR FROM C.
TITLE 2 GLUTAMICUM
SOURCE 2 ORGANISM_SCIENTIFIC: CORYNEBACTERIUM GLUTAMICUM;
SOURCE 3 ORGANISM_COMMON: BREVIBACTERIUM FLAVUM;
SOURCE 4 ORGANISM_TAXID: 1718;
KEYWDS GENE REGULATOR, HELIX-TURN-HELIX MOTIF, TRANSCRIPTIONAL
KEYWDS 2 ACTIVATOR, HUMAN PATHOGEN, TRANSCRIPTION ACTIVATOR
SCOP: none