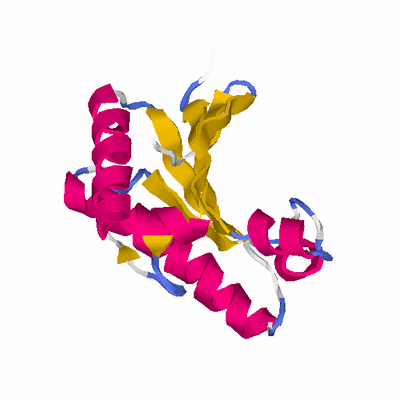
##########
# PROTEIN: Q2246_545_215.5wLII_11184_49 202 215 # Category_C1
# Length: 202
# Weight: 22436.36
# Similarity: UniProt_B6AMK1, BLAST_B6AMK1, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 61.39 M_00 2k1s_A 149 1 7e-36 25.000 124:128 (59-186:18-141)
# Evl 69.80 M_00 2k1s_A 149 1 2e-38 23.000 141:146 (41-186:1-141)
# Sid 40.10 M_01 1oap_A 109 1 7e-07 33.000 81:81 (77-157:7-87)
# Cov 69.80 M_00 2k1s_A 149 1 2e-38 23.000 141:146 (41-186:1-141)
# LaL 48.51 M_04 1r1m_A 164 1 4e-19 25.000 98:98 (68-165:8-105)
# OvN 34.65 M_07 1prx_A 224 2 0.059 22.000 70:70 (9-78:138-207)
# OvC 55.94 M_04 3cyp_C 138 20 2e-19 19.000 113:130 (83-202:12-137)
PDB: 2k1s PDBsum
HEADER LIPOPROTEIN 14-MAR-08 2K1S
TITLE SOLUTION NMR STRUCTURE OF THE FOLDED C-TERMINAL FRAGMENT OF
TITLE 2 YIAD FROM ESCHERICHIA COLI. NORTHEAST STRUCTURAL GENOMICS
TITLE 3 CONSORTIUM TARGET ER553.
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI K12;
SOURCE 3 ORGANISM_TAXID: 83333;
KEYWDS ABBABABAB, OMPA, LIPOPROTEIN, ALPHA BETA, INNER MEMBRANE,
KEYWDS 2 MEMBRANE, PALMITATE, TRANSMEMBRANE, STRUCTURAL GENOMICS,
KEYWDS 3 PSI-2, PROTEIN STRUCTURE INITIATIVE, NORTHEAST STRUCTURAL
KEYWDS 4 GENOMICS CONSORTIUM, NESG
SCOP: none
PDB: 1oap PDBsum
HEADER LIPOPROTEIN 20-JAN-03 1OAP
TITLE MAD STRUCTURE OF THE PERIPLASMIQUE DOMAIN OF THE
TITLE 2 ESCHERICHIA COLI PAL PROTEIN
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS PERIPLASMIC, PEPTIDOGLYCAN BINDING, TOL SYSTEM, OUTER
KEYWDS 2 MEMBRANE, SIGNAL, LIPOPROTEIN, COMPLETE PROTEOME
SCOP: d.79 Bacillus chorismate mutase-like
SCOP: d.79.7 OmpA-like
SCOP: d.79.7.1 OmpA-like
PDB: 1r1m PDBsum
HEADER MEMBRANE PROTEIN 24-SEP-03 1R1M
TITLE STRUCTURE OF THE OMPA-LIKE DOMAIN OF RMPM FROM NEISSERIA
TITLE 2 MENINGITIDIS
SOURCE 2 ORGANISM_SCIENTIFIC: NEISSERIA MENINGITIDIS;
SOURCE 3 ORGANISM_TAXID: 487;
KEYWDS MEMBRANE PROTEIN
SCOP: d.79 Bacillus chorismate mutase-like
SCOP: d.79.7 OmpA-like
SCOP: d.79.7.1 OmpA-like
PDB: 1prx PDBsum
HEADER ANTIOXIDANT 03-APR-98 1PRX
TITLE HORF6 A NOVEL HUMAN PEROXIDASE ENZYME
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
KEYWDS PEROXIREDOXIN, HORF6, HYDROGEN PEROXIDE, REDOX REGULATION,
KEYWDS 2 CELLULAR SIGNALING, ANTIOXIDANT
SCOP: c.47 Thioredoxin fold
SCOP: c.47.1 Thioredoxin-like
SCOP: c.47.1.10 Glutathione peroxidase-like
PDB: 3cyp PDBsum
HEADER MEMBRANE PROTEIN 26-APR-08 3CYP
TITLE THE CRYSTAL STRUCTURE OF THE C-TERMINAL DOMAIN OF
TITLE 2 HELICOBACTER PYLORI MOTB (RESIDUES 125-256).
SOURCE 2 ORGANISM_SCIENTIFIC: HELICOBACTER PYLORI;
SOURCE 3 ORGANISM_COMMON: CAMPYLOBACTER PYLORI;
KEYWDS HELICOBACTER PYLORI, BACTERIAL FLAGELLAR MOTOR,
KEYWDS 2 PEPTIDOGLYCAN BINDING, BACTERIAL FLAGELLUM, CHEMOTAXIS,
KEYWDS 3 FLAGELLAR ROTATION, INNER MEMBRANE, MEMBRANE, TRANSMEMBRANE,
KEYWDS 4 MEMBRANE PROTEIN
SCOP: none