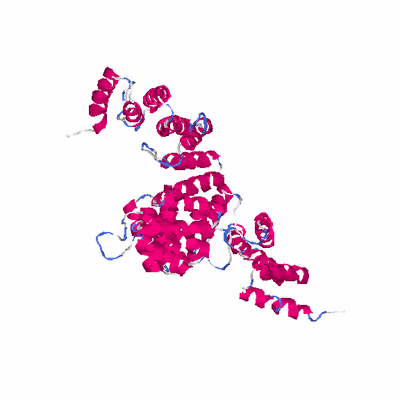
##########
# PROTEIN: Q2246_545_231.5wLII_11195_32.FR_1_700 700 231 # Category_C1
# Length: 700
# Weight: 80798.85
# Similarity: UniProt_B6AKK4, BLAST_B6AKK4, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 53.71 M_00 1w3b_A 388 2 9e-67 15.000 376:424 (217-640:13-388)
# Evl 54.86 M_00 1w3b_A 388 2 1e-76 13.000 384:432 (209-640:5-388)
# Sid 9.71 M_21 2avp_A 70 1 4e-15 23.000 68:69 (360-428:2-70)
# Cov 86.71 M_39 1xi4_A 1630 18 1e-08 8.000 607:725 (12-683:871-1530)
# LaL 18.71 M_30 1a17_A 166 1 2e-11 13.000 131:131 (437-567:13-143)
# OvN 33.71 M_30 1xnf_B 275 2 1e-08 13.000 236:267 (1-267:30-272)
# OvC 19.43 M_27 1p5q_A 336 3 5e-12 10.000 136:151 (564-699:121-271)
PDB: 1w3b PDBsum
HEADER TRANSFERASE 14-JUL-04 1W3B
TITLE THE SUPERHELICAL TPR DOMAIN OF O-LINKED GLCNAC TRANSFERASE
TITLE 2 REVEALS STRUCTURAL SIMILARITIES TO IMPORTIN ALPHA.
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 7 EC: 2.4.1.-;
KEYWDS OGT, GLCNAC, NUCLEOPORIN, O-LINKED GLYCOSYLATION, TPR
KEYWDS 2 REPEAT, PROTEIN BINDING, SIGNAL TRANSDUCTION, TRANSFERASE
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)
PDB: 2avp PDBsum
HEADER DE NOVO PROTEIN 30-AUG-05 2AVP
TITLE CRYSTAL STRUCTURE OF AN 8 REPEAT CONSENSUS TPR SUPERHELIX
KEYWDS TETRATRICOPEPTIDE REPEAT, TPR, CONSENSUS PROTEIN,
KEYWDS 2 SUPERHELIX, DE NOVO PROTEIN
SCOP: k.38 TPR domain-based design
SCOP: k.38.1 TPR domain-based design
SCOP: k.38.1.1 TPR domain-based design
PDB: 1xi4 PDBsum
HEADER ENDOCYTOSIS/EXOCYTOSIS 21-SEP-04 1XI4
TITLE CLATHRIN D6 COAT
SOURCE 2 ORGANISM_SCIENTIFIC: BOS TAURUS;
SOURCE 3 ORGANISM_COMMON: CATTLE;
SOURCE 4 ORGANISM_TAXID: 9913;
KEYWDS CLATHRIN, ALPHA-ZIG-ZAG, BETA-PROPELLER,
KEYWDS 2 ENDOCYTOSIS/EXOCYTOSIS COMPLEX
SCOP: i.23 clathrin assemblies
SCOP: i.23.1 clathrin assemblies
SCOP: i.23.1.1 clathrin assemblies
PDB: 1a17 PDBsum
HEADER HYDROLASE 23-DEC-97 1A17
TITLE TETRATRICOPEPTIDE REPEATS OF PROTEIN PHOSPHATASE 5
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 6 EC: 3.1.3.16;
KEYWDS HYDROLASE, PHOSPHATASE, PROTEIN-PROTEIN INTERACTIONS, TPR,
KEYWDS 2 SUPER-HELIX, X-RAY STRUCTURE
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)
PDB: 1xnf PDBsum
HEADER STRUCTURAL GENOMICS, UNKOWN FUNCTION 04-OCT-04 1XNF
TITLE CRYSTAL STRUCTURE OF E.COLI TPR-PROTEIN NLPI
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS NLPI, TPR, TETRATRICOPEPTIDE, LIPOPROTEIN, STRUCTURAL
KEYWDS 2 GENOMICS, UNKOWN FUNCTION
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)
PDB: 1p5q PDBsum
HEADER ISOMERASE 28-APR-03 1P5Q
TITLE CRYSTAL STRUCTURE OF FKBP52 C-TERMINAL DOMAIN
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 6 EC: 5.2.1.8;
KEYWDS ISOMERASE
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)
SCOP: d.26 FKBP-like
SCOP: d.26.1 FKBP-like
SCOP: d.26.1.1 FKBP immunophilin/proline isomerase