##########
# PROTEIN: Q2246_545_233.5wLII_11195_36 114 233 # Category_B
# Length: 114
# Weight: 12681.67
# Similarity: UniProt_B6AKK0, BLAST_B6AKK0, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 91.23 M_00 2pii_A 112 2 2e-39 26.000 104:110 (9-112:1-110)
# Evl 91.23 M_00 2pii_A 112 2 2e-39 26.000 104:110 (9-112:1-110)
# Sid 76.32 M_04 3bzq_A 114 1 3e-36 27.000 87:108 (9-110:3-110)
# Cov 92.98 M_10 2j9c_A 119 17 1e-34 24.000 106:112 (7-112:1-112)
# LaL 35.09 M_18 3bld_A 386 2 0.016 12.000 40:40 (26-65:216-255)
# OvN 83.33 M_13 2o66_B 135 8 1e-32 22.000 95:118 (2-112:6-123)
# OvC 50.00 M_25 2pq4_A 90 1 0.096 20.000 57:62 (53-114:34-90)
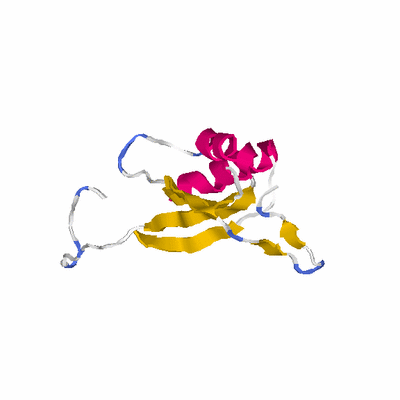
PDB: 2pii PDBsum
HEADER SIGNAL TRANSDUCTION PROTEIN 02-MAY-95 2PII
TITLE PII, GLNB PRODUCT
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
KEYWDS SIGNAL TRANSDUCTION PROTEIN
SCOP: d.58 Ferredoxin-like
SCOP: d.58.5 GlnB-like
SCOP: d.58.5.1 Prokaryotic signal transducing protein
PDB: 3bzq PDBsum
HEADER SIGNALING PROTEIN/TRANSCRIPTION 18-JAN-08 3BZQ
TITLE HIGH RESOLUTION CRYSTAL STRUCTURE OF NITROGEN REGULATORY
TITLE 2 PROTEIN (RV2919C) OF MYCOBACTERIUM TUBERCULOSIS
SOURCE 2 ORGANISM_SCIENTIFIC: MYCOBACTERIUM TUBERCULOSIS H37RV;
SOURCE 3 ORGANISM_TAXID: 83332;
KEYWDS P-II, GLNB, GLNK, MYCOBACTERIUM TUBERCULOSIS, SIGNAL
KEYWDS 2 TRANSDUCTORY PROTEIN, NUCLEOTIDE-BINDING, TRANSCRIPTION,
KEYWDS 3 TRANSCRIPTION REGULATION, SIGNAL TRANSDUCTION, NITROGEN
KEYWDS 4 REGULATORY, SIGNALING PROTEIN/TRANSCRIPTION COMPLEX,
KEYWDS 5 STRUCTURAL GENOMICS, TB STRUCTURAL GENOMICS CONSORTIUM,
KEYWDS 6 TBSGC
SCOP: none
PDB: 2j9c PDBsum
HEADER MEMBRANE TRANSPORT 07-NOV-06 2J9C
TITLE STRUCTURE OF GLNK1 WITH BOUND EFFECTORS INDICATES
TITLE 2 REGULATORY MECHANISM FOR AMMONIA UPTAKE
SOURCE 2 ORGANISM_SCIENTIFIC: METHANOCOCCUS JANNASCHII;
SOURCE 3 ORGANISM_TAXID: 2190;
KEYWDS EM SINGLE PARTICLE, NITROGEN METABOLISM, SIGNALLING,
KEYWDS 2 TRANSCRIPTION, MEMBRANE TRANSPORT, HYPOTHETICAL PROTEIN,
KEYWDS 3 TRANSCRIPTION REGULATION
SCOP: none
PDB: 3bld PDBsum
HEADER TRANSFERASE 11-DEC-07 3BLD
TITLE TRNA GUANINE TRANSGLYCOSYLASE V233G MUTANT PREQ1 COMPLEX
TITLE 2 STRUCTURE
SOURCE 2 ORGANISM_SCIENTIFIC: ZYMOMONAS MOBILIS;
SOURCE 3 ORGANISM_TAXID: 542;
COMPND 6 EC: 2.4.2.29;
KEYWDS TGT, PREQ1, GLYCOSYLTRANSFERASE, METAL-BINDING, QUEUOSINE
KEYWDS 2 BIOSYNTHESIS, TRANSFERASE, TRNA PROCESSING, ZINC
SCOP: none
PDB: 2o66 PDBsum
HEADER BIOSYNTHETIC PROTEIN 06-DEC-06 2O66
TITLE CRYSTAL STRUCTURE OF ARABIDOPSIS THALIANA PII BOUND TO
TITLE 2 CITRATE
SOURCE 2 ORGANISM_SCIENTIFIC: ARABIDOPSIS THALIANA;
SOURCE 3 ORGANISM_COMMON: THALE CRESS;
SOURCE 4 ORGANISM_TAXID: 3702;
KEYWDS REGULATION OF NITROGEN AND CARBON METABOLISM, BIOSYNTHETIC
KEYWDS 2 PROTEIN
SCOP: none
PDB: 2pq4 PDBsum
HEADER CHAPERONE/OXIDOREDUCTASE 01-MAY-07 2PQ4
TITLE NMR SOLUTION STRUCTURE OF NAPD IN COMPLEX WITH NAPA1-35
TITLE 2 SIGNAL PEPTIDE
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 10 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
COMPND 9 EC: 1.7.99.4;
KEYWDS NAPD/NAPA1-35, MIXED BETA-ALPHA SANDWICH STRUCTURE, PROTEIN-
KEYWDS 2 PEPTIDE COMPLEX, ALPHA-HELIX, STRUCTURAL GENOMICS, PSI,
KEYWDS 3 PROTEIN STRUCTURE INITIATIVE, MONTREAL-KINGSTON BACTERIAL
KEYWDS 4 STRUCTURAL GENOMICS INITIATIVE, BSGI,
KEYWDS 5 CHAPERONE/OXIDOREDUCTASE COMPLEX
SCOP: none