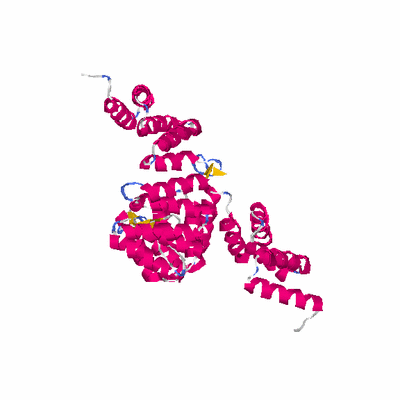
##########
# PROTEIN: Q2246_545_238.5wLII_11212_44 600 238 # Category_C1
# Length: 600
# Weight: 67640.28
# Similarity: UniProt_B6APK8, BLAST_B6APK8, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 59.33 M_00 1w3b_A 388 2 1e-58 15.000 356:394 (32-422:30-388)
# Evl 62.17 M_00 1w3b_A 388 2 2e-69 8.000 373:388 (208-588:1-380)
# Sid 11.33 M_16 2avp_A 70 1 7e-17 23.000 68:68 (305-372:3-70)
# Cov 76.00 M_08 2gw1_A 514 2 2e-23 11.000 456:497 (37-528:8-495)
# LaL 22.67 M_05 2fo7_A 136 2 2e-32 17.000 136:136 (305-440:1-136)
# OvN 69.67 M_07 2gw1_A 514 2 1e-24 10.000 418:475 (15-470:19-479)
# OvC 56.17 M_38 1xi4_A 1630 18 6e-09 9.000 337:385 (220-596:1032-1376)
PDB: 1w3b PDBsum
HEADER TRANSFERASE 14-JUL-04 1W3B
TITLE THE SUPERHELICAL TPR DOMAIN OF O-LINKED GLCNAC TRANSFERASE
TITLE 2 REVEALS STRUCTURAL SIMILARITIES TO IMPORTIN ALPHA.
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 7 EC: 2.4.1.-;
KEYWDS OGT, GLCNAC, NUCLEOPORIN, O-LINKED GLYCOSYLATION, TPR
KEYWDS 2 REPEAT, PROTEIN BINDING, SIGNAL TRANSDUCTION, TRANSFERASE
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)
PDB: 2avp PDBsum
HEADER DE NOVO PROTEIN 30-AUG-05 2AVP
TITLE CRYSTAL STRUCTURE OF AN 8 REPEAT CONSENSUS TPR SUPERHELIX
KEYWDS TETRATRICOPEPTIDE REPEAT, TPR, CONSENSUS PROTEIN,
KEYWDS 2 SUPERHELIX, DE NOVO PROTEIN
SCOP: k.38 TPR domain-based design
SCOP: k.38.1 TPR domain-based design
SCOP: k.38.1.1 TPR domain-based design
PDB: 2gw1 PDBsum
HEADER PROTEIN TRANSPORT 03-MAY-06 2GW1
TITLE CRYSTAL STRUCTURE OF THE YEAST TOM70
SOURCE 2 ORGANISM_SCIENTIFIC: SACCHAROMYCES CEREVISIAE;
SOURCE 3 ORGANISM_COMMON: BAKER'S YEAST;
SOURCE 4 ORGANISM_TAXID: 4932;
KEYWDS TPR, PROTEIN TRANSPORT
SCOP: none
PDB: 2fo7 PDBsum
HEADER DE NOVO PROTEIN 12-JAN-06 2FO7
TITLE CRYSTAL STRUCTURE OF AN 8 REPEAT CONSENSUS TPR SUPERHELIX
TITLE 2 (TRIGONAL CRYSTAL FORM)
KEYWDS TETRATRICOPEPTIDE REPEAT, TPR, CONSENSUS PROTEIN,
KEYWDS 2 SUPERHELIX, DE NOVO PROTEIN
SCOP: k.38 TPR domain-based design
SCOP: k.38.1 TPR domain-based design
SCOP: k.38.1.1 TPR domain-based design
PDB: 1xi4 PDBsum
HEADER ENDOCYTOSIS/EXOCYTOSIS 21-SEP-04 1XI4
TITLE CLATHRIN D6 COAT
SOURCE 2 ORGANISM_SCIENTIFIC: BOS TAURUS;
SOURCE 3 ORGANISM_COMMON: CATTLE;
SOURCE 4 ORGANISM_TAXID: 9913;
KEYWDS CLATHRIN, ALPHA-ZIG-ZAG, BETA-PROPELLER,
KEYWDS 2 ENDOCYTOSIS/EXOCYTOSIS COMPLEX
SCOP: i.23 clathrin assemblies
SCOP: i.23.1 clathrin assemblies
SCOP: i.23.1.1 clathrin assemblies