##########
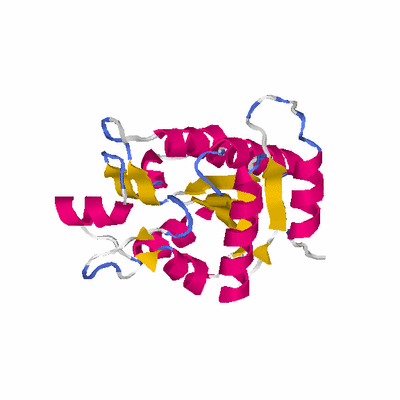
# PROTEIN: Q2246_545_245.5wLII_11212_69 237 245 # Category_B
# Length: 237
# Weight: 25618.63
# Similarity: UniProt_B6APN2, BLAST_B6APN2, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 80.59 M_00 1dts_A 221 1 2e-47 23.000 191:202 (29-225:1-197)
# Evl 80.59 M_00 1dts_A 221 1 2e-47 23.000 191:202 (29-225:1-197)
# Sid 15.61 M_04 z084 222 0 0.003 54.000 37:37 (34-70:6-42)
# Cov 85.23 M_02 z026 240 0 1e-41 22.000 202:218 (29-235:1-214)
# LaL 20.25 M_16 3bfv_A 271 2 0.002 14.000 48:48 (15-62:67-114)
# OvN 81.01 M_02 3cio_A 299 2 2e-39 13.000 192:215 (1-205:75-276)
# OvC 80.17 M_03 z08w 262 0 8e-38 12.000 190:232 (29-236:1-224)
PDB: 1dts PDBsum
HEADER CYCLO-LIGASE 28-MAR-95 1DTS
TITLE CRYSTAL STRUCTURE OF AN ATP DEPENDENT CARBOXYLASE,
TITLE 2 DETHIOBIOTIN SYNTHASE, AT 1.65 ANGSTROMS RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562
COMPND 4 EC: 6.3.3.3;
KEYWDS CYCLO-LIGASE
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.10 Nitrogenase iron protein-like
PDB: 1byi PDBsum
HEADER LIGASE 15-OCT-98 1BYI
TITLE STRUCTURE OF APO-DETHIOBIOTIN SYNTHASE AT 0.97 ANGSTROMS
TITLE 2 RESOLUTION
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562
COMPND 4 EC: 6.3.3.3
KEYWDS BIOTIN SYNTHESIS, CYCLO-LIGASE, LIGASE
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.10 Nitrogenase iron protein-like
PDB: 3bfv PDBsum
HEADER TRANSFERASE 23-NOV-07 3BFV
TITLE CRYSTAL STRUCTURE OF THE CHIMERICAL PROTEIN CAPAB
SOURCE 2 ORGANISM_SCIENTIFIC: STAPHYLOCOCCUS AUREUS;
KEYWDS CHIMERICAL PROTEIN, P-LOOP PROTEIN, CAPSULE
KEYWDS 2 BIOGENESIS/DEGRADATION, EXOPOLYSACCHARIDE SYNTHESIS,
KEYWDS 3 MEMBRANE, TRANSMEMBRANE, VIRULENCE, TRANSFERASE
SCOP: none
PDB: 3cio PDBsum
HEADER SIGNALING PROTEIN, TRANSFERASE 11-MAR-08 3CIO
TITLE THE KINASE DOMAIN OF ESCHERICHIA COLI TYROSINE KINASE ETK
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
COMPND 6 EC: 2.7.10.2;
KEYWDS ETK, WZC, ESCHERICHIA COLI TYROSINE KINASE DOMAIN,
KEYWDS 2 SIGNALING PROTEIN, TRANSFERASE, INNER MEMBRANE, MEMBRANE,
KEYWDS 3 PHOSPHOPROTEIN, TRANSMEMBRANE, TYROSINE-PROTEIN KINASE
SCOP: none
PDB: 2bek PDBsum
HEADER CHROMOSOME SEGREGATION 25-NOV-04 2BEK
TITLE STRUCTURE OF THE BACTERIAL CHROMOSOME SEGREGATION PROTEIN
TITLE 2 SOJ
SOURCE 2 ORGANISM_SCIENTIFIC: THERMUS THERMOPHILUS;
SOURCE 3 ORGANISM_TAXID: 262724;
KEYWDS SOJ, ATPASE, BACTERIAL, CHROMOSOME SEGREGATION
SCOP: none