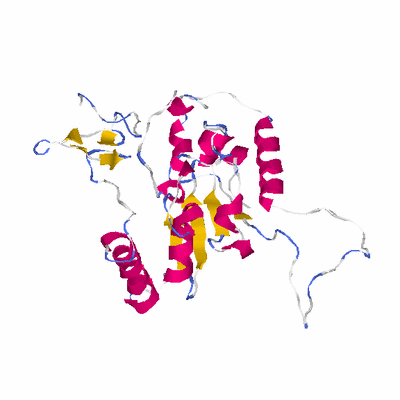
##########
# PROTEIN: Q2246_545_248.5wLII_11212_72 262 248 # Category_B
# Length: 262
# Weight: 29433.55
# Similarity: UniProt_B6APN5, BLAST_B6APN5, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 79.77 M_01 1m53_A 570 1 7e-53 20.000 209:258 (2-253:147-361)
# Evl 90.46 M_00 1pjr_A 724 3 2e-62 11.000 237:269 (6-262:51-300)
# Sid 51.15 M_00 1m53_A 570 1 0.004 27.000 134:147 (54-195:190-328)
# Cov 90.84 M_01 2pjr_F 548 2 2e-60 11.000 238:269 (6-262:51-300)
# LaL 42.37 M_06 2iue_A 212 1 2e-06 10.000 111:111 (28-138:3-113)
# OvN 72.52 M_11 1m7x_B 617 4 1e-11 10.000 190:269 (1-228:229-478)
# OvC 90.46 M_00 1pjr_A 724 3 2e-62 11.000 237:269 (6-262:51-300)
PDB: 1m53 PDBsum
HEADER ISOMERASE 08-JUL-02 1M53
TITLE CRYSTAL STRUCTURE OF ISOMALTULOSE SYNTHASE (PALI) FROM
TITLE 2 KLEBSIELLA SP. LX3
SOURCE 2 ORGANISM_SCIENTIFIC: KLEBSIELLA SP. LX3;
SOURCE 3 ORGANISM_TAXID: 167956;
COMPND 5 EC: 5.4.99.11;
KEYWDS KLEBSIELLA SP. LX3, ISOMALTULOSE SYNTHASE, SUCROSE
KEYWDS 2 ISOMERIZATION, ISOMERASE
SCOP: b.71 Glycosyl hydrolase domain
SCOP: b.71.1 Glycosyl hydrolase domain
SCOP: b.71.1.1 alpha-Amylases, C-terminal beta-sheet domain
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.8 (Trans)glycosidases
SCOP: c.1.8.1 Amylase, catalytic domain
PDB: 1pjr PDBsum
HEADER HELICASE 11-OCT-96 1PJR
TITLE STRUCTURE OF DNA HELICASE
SOURCE 2 ORGANISM_SCIENTIFIC: GEOBACILLUS STEAROTHERMOPHILUS;
SOURCE 3 ORGANISM_TAXID: 1422;
KEYWDS DNA REPAIR, DNA REPLICATION, SOS RESPONSE, HELICASE, ATP-
KEYWDS 2 BINDING, DNA-BINDING
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.19 Tandem AAA-ATPase domain
PDB: 2pjr PDBsum
HEADER HYDROLASE/DNA 12-MAR-99 2PJR
TITLE HELICASE PRODUCT COMPLEX
SOURCE 8 ORGANISM_SCIENTIFIC: GEOBACILLUS STEAROTHERMOPHILUS;
SOURCE 9 ORGANISM_TAXID: 1422;
SOURCE 13 ORGANISM_SCIENTIFIC: GEOBACILLUS STEAROTHERMOPHILUS;
COMPND 18 EC: 3.6.1.-;
COMPND 25 EC: 3.6.1.-;
KEYWDS HELICASE PCRA, HYDROLASE, DNA, PRODUCT COMPLEX,
KEYWDS 2 HYDROLASE/DNA COMPLEX
SCOP: c.37 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1 P-loop containing nucleoside triphosphate hydrolases
SCOP: c.37.1.19 Tandem AAA-ATPase domain
PDB: 2iue PDBsum
HEADER MEMBRANE PROTEIN 02-JUN-06 2IUE
TITLE PACTOLUS I-DOMAIN: FUNCTIONAL SWITCHING OF THE ROSSMANN
TITLE 2 FOLD
SOURCE 2 ORGANISM_SCIENTIFIC: MUS MUSCULUS;
SOURCE 3 ORGANISM_COMMON: MOUSE;
SOURCE 4 ORGANISM_TAXID: 10090;
KEYWDS MEMBRANE PROTEIN, CD, ITC, LIMBS, MIDAS, ADMIDAS, MEMBRANE,
KEYWDS 2 INTEGRIN, TITRATION, ROSSMAN FOLD, CELL ADHESION,
KEYWDS 3 TRANSMEMBRANE
SCOP: none
PDB: 1m7x PDBsum
HEADER TRANSFERASE 23-JUL-02 1M7X
TITLE THE X-RAY CRYSTALLOGRAPHIC STRUCTURE OF BRANCHING ENZYME
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 6 EC: 2.4.1.18;
KEYWDS ALPHA/BETA BARREL, BETA SANDWICH, TRANSFERASE
SCOP: b.1 Immunoglobulin-like beta-sandwich
SCOP: b.1.18 E set domains
SCOP: b.1.18.2 E-set domains of sugar-utilizing enzymes
SCOP: b.71 Glycosyl hydrolase domain
SCOP: b.71.1 Glycosyl hydrolase domain
SCOP: b.71.1.1 alpha-Amylases, C-terminal beta-sheet domain
SCOP: c.1 TIM beta/alpha-barrel
SCOP: c.1.8 (Trans)glycosidases
SCOP: c.1.8.1 Amylase, catalytic domain