##########
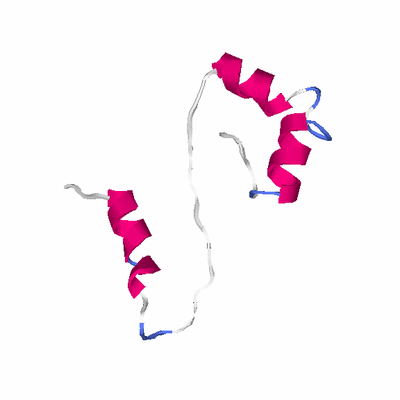
# PROTEIN: Q2246_545_26.5wLII_10665_6 76 26 # Category_B
# Length: 76
# Weight: 8201.70
# Similarity: UniProt_A3ER41, BLAST_A3ER41, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 76.32 M_00 2jzx_A 160 1 4e-09 27.000 58:62 (7-67:65-123)
# Evl 18.42 M_00 3cdi_A 723 1 5e-17 25.000 14:64 (3-66:535-593)
# Sid 39.47 M_01 1we8_A 104 1 0.038 38.000 30:31 (26-55:10-40)
# Cov 97.37 M_00 2pt7_G 152 2 1e-08 20.000 74:75 (1-74:4-78)
# LaL 76.32 M_09 2cxc_A 144 2 0.052 25.000 58:58 (5-62:78-135)
# OvN 97.37 M_00 2pt7_G 152 2 1e-08 20.000 74:75 (1-74:4-78)
# OvC 89.47 M_00 1vs5_C 233 2 2e-12 20.000 68:74 (3-76:38-105)
PDB: 2jzx PDBsum
HEADER RNA BINDING PROTEIN 21-JAN-08 2JZX
TITLE PCBP2 KH1-KH2 DOMAINS
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
KEYWDS PCBP2, KH DOMAINS, RNA BINDING, CYTOPLASM, DNA-BINDING,
KEYWDS 2 NUCLEUS, PHOSPHOPROTEIN, RIBONUCLEOPROTEIN, RNA-BINDING,
KEYWDS 3 RNA BINDING PROTEIN
SCOP: none
PDB: 3cdi PDBsum
HEADER TRANSFERASE 27-FEB-08 3CDI
TITLE CRYSTAL STRUCTURE OF E. COLI PNPASE
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
COMPND 5 EC: 2.7.7.8;
KEYWDS POLYNUCLEOTIDE PHOSPHORYLASE, MRNA TURNOVER, RNASE, RNA
KEYWDS 2 DEGRADATION, CRYSTAL STRUCTURE, KINASE, TRANSFERASE
SCOP: none
PDB: 1we8 PDBsum
HEADER RNA BINDING PROTEIN 24-MAY-04 1WE8
TITLE SOLUTION STRUCTURE OF KH DOMAIN IN PROTEIN BAB28342
SOURCE 2 ORGANISM_SCIENTIFIC: MUS MUSCULUS;
SOURCE 3 ORGANISM_COMMON: HOUSE MOUSE;
SOURCE 4 ORGANISM_TAXID: 10090;
KEYWDS NMR, STRUCTURAL GENOMICS, KH DOMAIN, RIKEN STRUCTURAL
KEYWDS 2 GENOMICS/PROTEOMICS INITIATIVE, RSGI, RNA BINDING PROTEIN
SCOP: d.51 Eukaryotic type KH-domain (KH-domain type I)
SCOP: d.51.1 Eukaryotic type KH-domain (KH-domain type I)
SCOP: d.51.1.1 Eukaryotic type KH-domain (KH-domain type I)
PDB: 2pt7 PDBsum
HEADER HYDROLASE/PROTEIN BINDING 08-MAY-07 2PT7
TITLE CRYSTAL STRUCTURE OF CAG VIRB11 (HP0525) AND AN INHIBITORY
TITLE 2 PROTEIN (HP1451)
SOURCE 2 ORGANISM_SCIENTIFIC: HELICOBACTER PYLORI 26695;
SOURCE 3 ORGANISM_TAXID: 85962;
SOURCE 12 ORGANISM_SCIENTIFIC: HELICOBACTER PYLORI 26695;
KEYWDS ATPASE, PROTEIN-PROTEIN COMPLEX, TYPE IV SECRETION,
KEYWDS 2 HYDROLASE/PROTEIN BINDING COMPLEX
SCOP: none
PDB: 2cxc PDBsum
HEADER TRANSCRIPTION 28-JUN-05 2CXC
TITLE CRYSTAL STRUCTURE OF ARCHAEAL TRANSCRIPTION TERMINATION
TITLE 2 FACTOR NUSA
SOURCE 2 ORGANISM_SCIENTIFIC: AEROPYRUM PERNIX K1;
SOURCE 3 ORGANISM_TAXID: 272557;
KEYWDS TRANSCRIPTION TERMINATION, RNA BINDING PROTEIN, ARCHAEAL
KEYWDS 2 NUSA, KH DOMAIN, STRUCTURAL GENOMICS, NPPSFA, NATIONAL
KEYWDS 3 PROJECT ON PROTEIN STRUCTURAL AND FUNCTIONAL ANALYSES,
KEYWDS 4 RIKEN STRUCTURAL GENOMICS/PROTEOMICS INITIATIVE, RSGI
SCOP: none
PDB: 1vs5 PDBsum
HEADER RIBOSOME 04-AUG-06 1VS5
TITLE CRYSTAL STRUCTURE OF THE BACTERIAL RIBOSOME FROM
TITLE 2 ESCHERICHIA COLI IN COMPLEX WITH THE ANTIBIOTIC KASUGAMYIN
TITLE 3 AT 3.5A RESOLUTION. THIS FILE CONTAINS THE 30S SUBUNIT OF
TITLE 4 ONE 70S RIBOSOME. THE ENTIRE CRYSTAL STRUCTURE CONTAINS
TITLE 5 TWO 70S RIBOSOMES AND IS DESCRIBED IN REMARK 400.
SOURCE 2 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
SOURCE 3 ORGANISM_TAXID: 562;
SOURCE 6 ORGANISM_SCIENTIFIC: ESCHERICHIA COLI;
KEYWDS RIBOSOME, KASUGAMYCIN
SCOP: d.140 Ribosomal protein S8
SCOP: d.140.1 Ribosomal protein S8
SCOP: d.140.1.1 Ribosomal protein S8