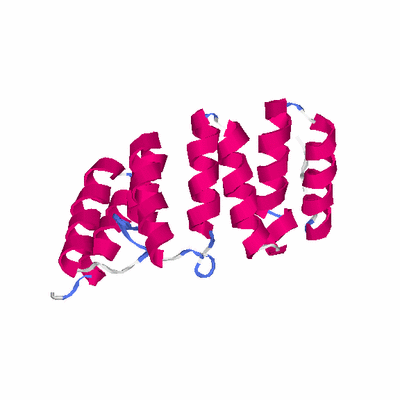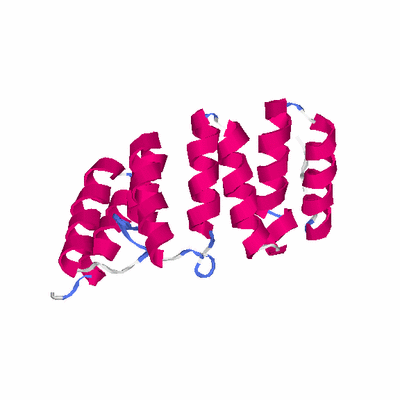##########
# PROTEIN: Q2246_545_268.5wLII_11233_37 264 268 # Category_C1
# Length: 264
# Weight: 28996.28
# Similarity: UniProt_A3EW63, BLAST_A3EW63, PDB_templates_INFO
# Best Pcover Model PDB N_AA SISC E-val Seq_ID LAL Overlap
# Cat 51.52 M_00 2fo7_A 136 2 2e-40 20.000 136:146 (84-229:1-136)
# Evl 50.38 M_00 2fo7_A 136 2 6e-41 19.000 133:142 (87-228:3-135)
# Sid 25.76 M_05 2avp_A 70 1 1e-20 25.000 68:70 (160-229:3-70)
# Cov 74.24 M_01 1w3b_A 388 2 1e-39 10.000 196:210 (35-242:188-385)
# LaL 30.68 M_04 1na3_A 91 2 1e-22 24.000 81:83 (160-242:9-89)
# OvN 64.39 M_23 1kt1_A 457 1 3e-13 14.000 170:211 (32-242:228-397)
# OvC 61.74 M_12 2gw1_A 514 2 2e-17 13.000 163:179 (77-248:261-430)
PDB: 2fo7 PDBsum
HEADER DE NOVO PROTEIN 12-JAN-06 2FO7
TITLE CRYSTAL STRUCTURE OF AN 8 REPEAT CONSENSUS TPR SUPERHELIX
TITLE 2 (TRIGONAL CRYSTAL FORM)
KEYWDS TETRATRICOPEPTIDE REPEAT, TPR, CONSENSUS PROTEIN,
KEYWDS 2 SUPERHELIX, DE NOVO PROTEIN
SCOP: k.38 TPR domain-based design
SCOP: k.38.1 TPR domain-based design
SCOP: k.38.1.1 TPR domain-based design
PDB: 2avp PDBsum
HEADER DE NOVO PROTEIN 30-AUG-05 2AVP
TITLE CRYSTAL STRUCTURE OF AN 8 REPEAT CONSENSUS TPR SUPERHELIX
KEYWDS TETRATRICOPEPTIDE REPEAT, TPR, CONSENSUS PROTEIN,
KEYWDS 2 SUPERHELIX, DE NOVO PROTEIN
SCOP: k.38 TPR domain-based design
SCOP: k.38.1 TPR domain-based design
SCOP: k.38.1.1 TPR domain-based design
PDB: 1w3b PDBsum
HEADER TRANSFERASE 14-JUL-04 1W3B
TITLE THE SUPERHELICAL TPR DOMAIN OF O-LINKED GLCNAC TRANSFERASE
TITLE 2 REVEALS STRUCTURAL SIMILARITIES TO IMPORTIN ALPHA.
SOURCE 2 ORGANISM_SCIENTIFIC: HOMO SAPIENS;
SOURCE 3 ORGANISM_COMMON: HUMAN;
SOURCE 4 ORGANISM_TAXID: 9606;
COMPND 7 EC: 2.4.1.-;
KEYWDS OGT, GLCNAC, NUCLEOPORIN, O-LINKED GLYCOSYLATION, TPR
KEYWDS 2 REPEAT, PROTEIN BINDING, SIGNAL TRANSDUCTION, TRANSFERASE
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)
PDB: 1na3 PDBsum
HEADER DE NOVO PROTEIN 26-NOV-02 1NA3
TITLE DESIGN OF STABLE ALPHA-HELICAL ARRAYS FROM AN IDEALIZED TPR
TITLE 2 MOTIF
SOURCE 2 ORGANISM_SCIENTIFIC: UNIDENTIFIED;
SOURCE 3 ORGANISM_TAXID: 32644;
KEYWDS DESIGN, TPR, DE NOVO PROTEIN
SCOP: k.38 TPR domain-based design
SCOP: k.38.1 TPR domain-based design
SCOP: k.38.1.1 TPR domain-based design
PDB: 1kt1 PDBsum
HEADER ISOMERASE 14-JAN-02 1KT1
TITLE STRUCTURE OF THE LARGE FKBP-LIKE PROTEIN, FKBP51, INVOLVED
TITLE 2 IN STEROID RECEPTOR COMPLEXES
SOURCE 2 ORGANISM_SCIENTIFIC: SAIMIRI BOLIVIENSIS;
SOURCE 3 ORGANISM_COMMON: BOLIVIAN SQUIRREL MONKEY;
SOURCE 4 ORGANISM_TAXID: 27679;
COMPND 4 EC: 5.2.1.8;
KEYWDS FKBP-LIKE PPIASE, TPR REPEATS, ISOMERASE
SCOP: a.118 alpha-alpha superhelix
SCOP: a.118.8 TPR-like
SCOP: a.118.8.1 Tetratricopeptide repeat (TPR)
SCOP: d.26 FKBP-like
SCOP: d.26.1 FKBP-like
SCOP: d.26.1.1 FKBP immunophilin/proline isomerase
PDB: 2gw1 PDBsum
HEADER PROTEIN TRANSPORT 03-MAY-06 2GW1
TITLE CRYSTAL STRUCTURE OF THE YEAST TOM70
SOURCE 2 ORGANISM_SCIENTIFIC: SACCHAROMYCES CEREVISIAE;
SOURCE 3 ORGANISM_COMMON: BAKER'S YEAST;
SOURCE 4 ORGANISM_TAXID: 4932;
KEYWDS TPR, PROTEIN TRANSPORT
SCOP: none